Metabolic Network Flux Balance Analysis of the Microgravity Influence on the Growth of Arabidopsis Thaliana
-
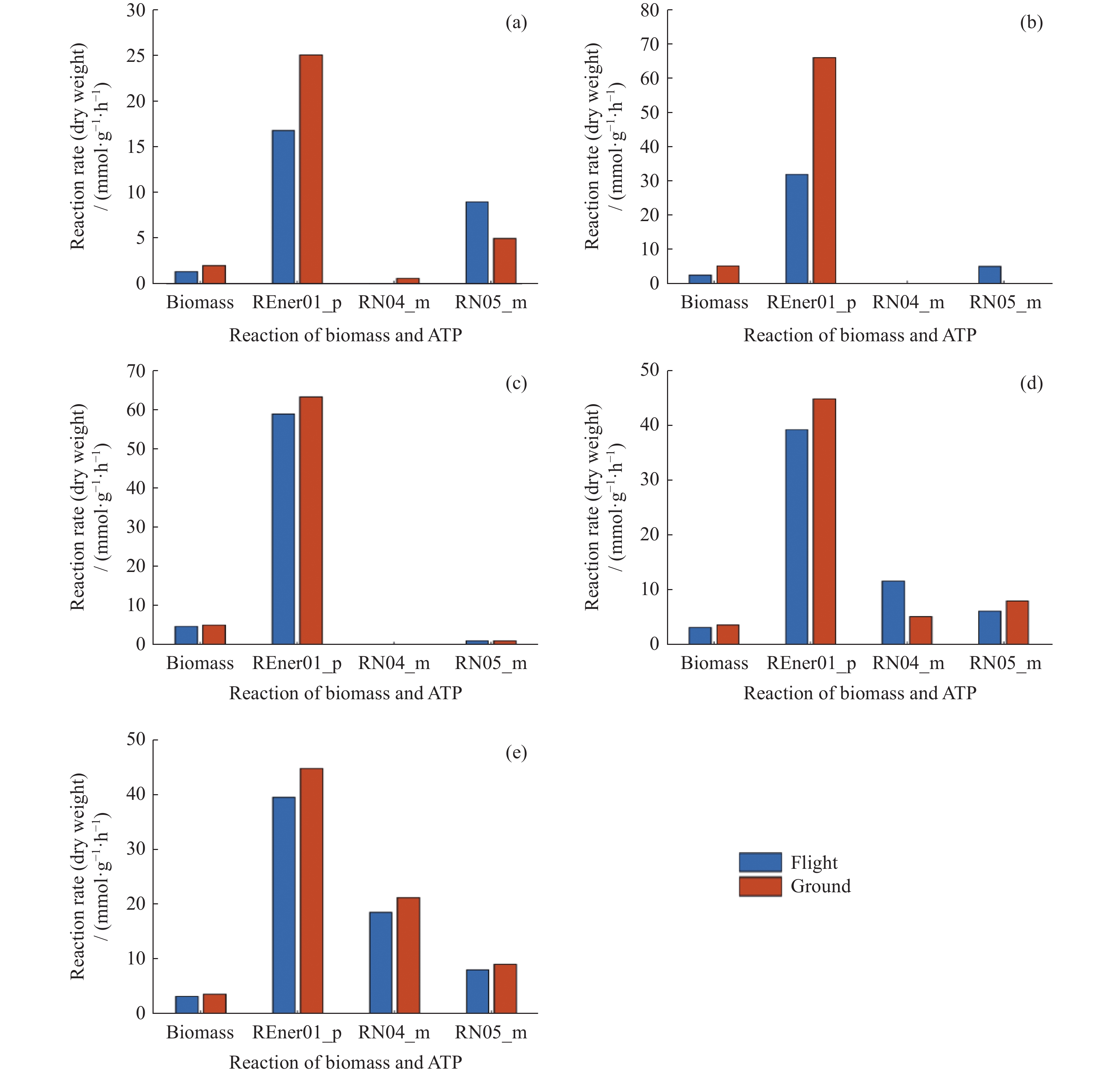
摘要: 微重力作为典型的空间环境因素,对植物生长发育的影响机制是空间生命科学的研究热点。微重力环境直接或间接影响植物代谢,并引起许多生理适应。 随着系统生物学的发展,代谢网络模型使微重力环境下的植物代谢建模成为可能。采用流平衡分析方法对模式植物拟南芥不同组织的代谢网络进行分析,研究微重力对拟南芥生长发育的影响机制。通过比较空间与地面条件下拟南芥的生物质产量,发现空间条件下拟南芥黄化幼苗、幼苗、芽、根、下胚轴的生物量分别下降了33.00%,51.52%,6.89%,12.53%,11.70%,与空间环境下拟南芥的长势变化趋势一致。代谢通路富集分析发现,微重力使得拟南芥的碳固定等通路下调,而磷酸戊糖途径上调,初步解析了微重力对拟南芥生长发育的影响机制,也验证了流平衡方法用于微重力生物学效应研究中的可行性。Abstract: As a typical space environmental factor, the influence mechanism of microgravity on plants is a research hotspot in space life science. The microgravity environment directly or indirectly affects plant metabolism and induces many physiological adaptations. With the development of systems biology, the metabolic network model enables the modeling of plant metabolism in microgravity. In this study, the flux balance analysis method was used to study the metabolic networks of different tissues of the model plant Arabidopsis thaliana, and to explore the mechanism of the effect of microgravity on the growth and development of Arabidopsis thaliana. By comparing the biomass yield of Arabidopsis thaliana under space and ground conditions, it was found that the biomass of etiolated seedlings, seedlings, shoots, roots, and hypocotyls is decreased by 33.00%, 51.52%, 6.89%, 12.53%, and 11.70%, respectively, consistent with the growth trend of Arabidopsis thaliana in the space environment. Enrichment analysis of metabolic pathways showed that microgravity down-regulated Arabidopsis thaliana’s carbon fixation pathway, while up-regulated the pentose phosphate pathway. This result preliminarily analyzed the mechanism of microgravity influence on the growth and development of Arabidopsis thaliana and demonstrated the potential of the flux balance analysis method in the study of the biological effects of microgravity.
-
Key words:
- Microgravity /
- Metabolic network model /
- Flux balance analysis
-
表 1 拟南芥黄化幼苗下调反应对应基因的富集途径
Table 1. Enrichment pathways for genes corresponding to Arabidopsis thaliana etiolated seedlings down-regulation reactions
通路 KEGG通路ID p 值 FDR Glyoxylate and dicarboxylate metabolism ath00630 4.24×10–24 1.10×10–22 Carbon metabolism ath01200 1.88×10–16 2.44×10–15 Biosynthesis of antibiotics ath01130 2.20×10–11 1.91×10–10 Metabolic pathways ath01100 4.68×10–11 3.04×10–10 Biosynthesis of secondary metabolites ath01110 6.97×10–10 3.62×10–9 Carbon fixation in photosynthetic organisms ath00710 1.21×10–9 5.24×10–9 Pyruvate metabolism ath00620 6.00×10–9 2.23×10–8 Citrate cycle (TCA cycle) ath00020 2.12×10–8 6.88×10–8 Cysteine and methionine metabolism ath00270 5.04×10–7 1.46×10–6 One carbon pool by folate ath00670 2.79×10–6 7.24×10–6 表 2 拟南芥幼苗下调反应对应基因的富集途径
Table 2. Enrichment pathways for genes corresponding to Arabidopsis thaliana seedlings down-regulation reactions
通路 KEGG通路ID p 值 FDR Oxidative phosphorylation ath00190 3.16×10–25 9.49×10–24 Metabolic pathways ath01100 2.24×10–24 3.36×10–23 Biosynthesis of amino acids ath01230 7.08×10–24 7.08×10–23 Phenylalanine, tyrosine and tryptophan biosynthesis ath00400 3.60×10–16 2.70×10–15 Carbon fixation in photosynthetic organisms ath00710 1.82×10–15 1.09×10–14 Biosynthesis of secondary metabolites ath01110 1.96×10–12 9.82×10–12 Biosynthesis of antibiotics ath01130 1.12×10–10 4.79×10–10 Phagosome ath04145 1.81×10–10 6.78×10–10 Carbon metabolism ath01200 2.82×10–10 9.40×10–10 Nitrogen metabolism ath00910 5.95×10–9 1.79×10–8 表 3 拟南芥芽下调反应对应基因的富集途径
Table 3. Enrichment pathways for genes corresponding to Arabidopsis thaliana shoot down-regulation reactions
通路 KEGG通路ID p 值 FDR Carbon metabolism ath01200 8.48×10–44 1.36×10–42 Biosynthesis of antibiotics ath01130 5.95×10–40 4.76×10–39 Pentose phosphate pathway ath00030 2.01×10–35 1.07×10–34 Glycolysis / Gluconeogenesis ath00010 2.27×10–33 9.09×10–33 Carbon fixation in photosynthetic organisms ath00710 2.20×10–28 7.05×10–28 Biosynthesis of secondary metabolites ath01110 2.04×10–23 5.43×10–23 Metabolic pathways ath01100 3.75×10–16 8.58×10–16 Fructose and mannose metabolism ath00051 1.05×10–13 2.10×10–13 Nitrogen metabolism ath00910 3.26×10–13 5.79×10–13 Biosynthesis of amino acids ath01230 4.02×10–12 6.43×10–12 表 4 拟南芥根下调反应对应基因的富集途径
Table 4. Enrichment pathways for genes corresponding to Arabidopsis thaliana root down-regulation reactions
通路 KEGG通路ID p 值 FDR Carbon metabolism ath01200 3.50×10–35 4.20×10–34 Carbon fixation in photosynthetic organisms ath00710 1.42×10–34 8.53×10–34 Glycolysis / Gluconeogenesis ath00010 1.63×10–23 6.53×10–23 Biosynthesis of amino acids ath01230 1.16×10–22 3.48×10–22 Biosynthesis of antibiotics ath01130 2.89×10–20 6.94×10–20 Metabolic pathways ath01100 5.74×10–19 1.15×10–18 Pentose phosphate pathway ath00030 8.53×10–13 1.46×10–12 Fructose and mannose metabolism ath00051 4.30×10–12 6.45×10–12 Biosynthesis of secondary metabolites ath01110 3.44×10–11 4.58×10–11 Arginine biosynthesis ath00220 5.65×10–10 6.78×10–10 表 5 拟南芥下胚轴下调反应对应基因的富集途径
Table 5. Enrichment pathways for genes corresponding to Arabidopsis thaliana hypocotyl down-regulation reactions
通路 KEGG通路ID p 值 FDR Carbon fixation in photosynthetic organisms ath00710 1.05×10–30 1.48×10–29 Carbon metabolism ath01200 1.40×10–28 9.80×10–28 Glycolysis / Gluconeogenesis ath00010 3.94×10–23 1.84×10–22 Biosynthesis of antibiotics ath01130 1.33×10–20 4.66×10–20 Biosynthesis of secondary metabolites ath01110 1.25×10–19 3.49×10–19 Metabolic pathways ath01100 3.46×10–18 8.08×10–18 Fructose and mannose metabolism ath00051 2.05×10–15 4.10×10–15 Biosynthesis of amino acids ath01230 1.42×10–14 2.48×10–14 Pentose phosphate pathway ath00030 1.95×10–13 3.03×10–13 Arginine biosynthesis ath00220 7.01×10–10 9.82×10–10 表 6 拟南芥黄化幼苗上调反应对应基因的富集途径
Table 6. Enrichment pathways for genes corresponding to Arabidopsis thaliana etiolated seedlings up-regulation reactions
通路 KEGG通路ID p 值 FDR Carbon metabolism ath01200 3.78×10–37 4.16×10–36 Biosynthesis of antibiotics ath01130 9.37×10–22 5.15×10–21 Pentose phosphate pathway ath00030 4.01×10–18 1.47×10–17 Carbon fixation in photosynthetic organisms ath00710 6.44×10–18 1.77×10–17 Nitrogen metabolism ath00910 1.45×10–11 3.18×10–11 Biosynthesis of secondary metabolites ath01110 2.22×10–11 4.07×10–11 Biosynthesis of amino acids ath01230 9.14×10–11 1.44×10–10 Metabolic pathways ath01100 1.76×10–9 2.42×10–9 Glycolysis / Gluconeogenesis ath00010 3.36×10–7 4.11×10–7 Glutathione metabolism ath00480 7.11×10–6 7.82×10–6 表 7 拟南芥幼苗上调反应对应基因的富集途径
Table 7. Enrichment pathways for genes corresponding to Arabidopsis thaliana seedlings up-regulation reactions
通路 KEGG通路ID p 值 FDR Carbon metabolism ath01200 1.85×10–23 1.37×10–22 Pentose phosphate pathway ath00030 2.28×10–23 1.37×10–22 Biosynthesis of antibiotics ath01130 5.24×10–23 2.10×10–22 Biosynthesis of secondary metabolites ath01110 2.05×10–20 6.14×10–20 Metabolic pathways ath01100 3.38×10–15 8.12×10–15 Phenylalanine metabolism ath00360 1.27×10–8 2.55×10–8 Citrate cycle (TCA cycle) ath00020 2.06×10–8 3.53×10–8 Carbon fixation in photosynthetic organisms ath00710 9.38×10–6 1.41×10–5 Glycolysis / Gluconeogenesis ath00010 3.01×10–5 4.01×10–5 表 8 拟南芥下胚轴上调反应对应基因的富集途径
Table 8. Enrichment pathways for genes corresponding to Arabidopsis thaliana hypocotyl up-regulation reactions
通路 KEGG通路ID p 值 FDR Glyoxylate and dicarboxylate metabolism ath00630 1.12×10–21 1.23×10–20 Nitrogen metabolism ath00910 2.19×10–20 1.21×10–19 Carbon metabolism ath01200 7.92×10–9 2.91×10–8 Carbon fixation in photosynthetic organisms ath00710 5.85×10–8 1.61×10–7 Alanine, aspartate and glutamate metabolism ath00250 8.51×10–8 1.87×10–7 Pyruvate metabolism ath00620 2.79×10–7 5.12×10–7 One carbon pool by folate ath00670 4.16×10–7 6.53×10–7 Citrate cycle (TCA cycle) ath00020 5.85×10–7 8.04×10–7 Biosynthesis of antibiotics ath01130 8.07×10–7 9.86×10–7 Cysteine and methionine metabolism ath00270 1.27×10–5 1.40×10–5 表 9 拟南芥根上调反应对应基因的富集途径
Table 9. Enrichment pathways for genes corresponding to Arabidopsis thaliana root up-regulation reactions
通路 KEGG通路ID p 值 FDR Oxidative phosphorylation ath00190 3.63×10–32 1.02×10–30 Metabolic pathways ath01100 2.07×10–25 2.90×10–24 Biosynthesis of secondary metabolites ath01110 1.85×10–18 1.73×10–17 Biosynthesis of amino acids ath01230 3.81×10–15 2.67×10–14 Biosynthesis of antibiotics ath01130 6.91×10–15 3.87×10–14 Citrate cycle (TCA cycle) ath00020 1.77×10–14 8.24×10–14 Phenylalanine, tyrosine and tryptophan biosynthesis ath00400 5.69×10–13 2.27×10–12 2-Oxocarboxylic acid metabolism ath01210 5.91×10–11 2.07×10–10 Phagosome ath04145 6.69×10–11 2.08×10–10 Phenylalanine metabolism ath00360 2.09×10–10 5.84×10–10 表 10 拟南芥芽上调反应对应基因的富集途径
Table 10. Enrichment pathways for genes corresponding to Arabidopsis thaliana shoot up-regulation reactions
通路 KEGG通路ID p 值 FDR Oxidative phosphorylation ath00190 4.27×10–47 5.98×10–46 Phagosome ath04145 1.45×10–18 1.02×10–17 Metabolic pathways ath01100 5.92×10–12 2.76×10–11 Ascorbate and aldarate metabolism ath00053 1.25×10–4 4.37×10–4 One carbon pool by folate ath00670 0.001723 0.004825 -
[1] FU Y M, LI L Y, XIE B Z, et al. How to establish a bioregenerative life support system for long-term crewed missions to the moon or mars[J]. Astrobiology, 2016, 16(12): 925-936 doi: 10.1089/ast.2016.1477 [2] DE PASCALE S, ARENA C, ARONNE G, et al. Biology and crop production in Space environments: challenges and opportunities[J]. Life Sciences in Space Research, 2021, 29: 30-37 doi: 10.1016/j.lssr.2021.02.005 [3] SATHASIVAM M, HOSAMANI R, SWAMY B K, et al. Plant responses to real and simulated microgravity[J]. Life Sciences in Space Research, 2021, 28: 74-86 doi: 10.1016/j.lssr.2020.10.001 [4] SOLEIMANI M, GHANATI F, HAJEBRAHIMI Z, et al. Energy saving and improvement of metabolism of cultured tobacco cells upon exposure to 2-D clinorotation[J]. Journal of Plant Physiology, 2019, 234-235: 36-43 doi: 10.1016/j.jplph.2019.01.002 [5] ZHENG H Q, HAN F, LE J. Higher plants in space: microgravity perception, response, and adaptation[J]. Microgravity Science and Technology, 2015, 27(6): 377-386 doi: 10.1007/s12217-015-9428-y [6] ZHANG Y, WANG L H, XIE J Y, et al. Differential protein expression profiling of Arabidopsis thaliana callus under microgravity on board the Chinese SZ-8 spacecraft[J]. Planta, 2015, 241(2): 475-488 doi: 10.1007/s00425-014-2196-x [7] STRAUCH S M, GRIMM D, CORYDON T J, et al. Current knowledge about the impact of microgravity on the proteome[J]. Expert Review of Proteomics, 2019, 16(1): 5-16 doi: 10.1080/14789450.2019.1550362 [8] HEIRENDT L, ARRECKX S, PFAU T, et al. Creation and analysis of biochemical constraint-based models using the COBRA Toolbox v. 3.0[J]. Nature Protocols, 2019, 14(3): 639-702 doi: 10.1038/s41596-018-0098-2 [9] CHO J S, GU C D, HAN T H, et al. Reconstruction of context-specific genome-scale metabolic models using multiomics data to study metabolic rewiring[J]. Current Opinion in Systems Biology, 2019, 15: 1-11 doi: 10.1016/j.coisb.2019.02.009 [10] PAUL A L, ZUPANSKA A K, OSTROW D T, et al. Spaceflight transcriptomes: unique responses to a novel environment[J]. Astrobiology, 2012, 12(1): 40-56 doi: 10.1089/ast.2011.0696 [11] PAUL A L, ZUPANSKA A K, SCHULTZ E R, et al. Organ-specific remodeling of the Arabidopsis transcriptome in response to spaceflight[J]. BMC Plant Biology, 2013, 13: 112 doi: 10.1186/1471-2229-13-112 [12] IRIZARRY R A, HOBBS B, COLLIN F, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data[J]. Biostatistics, 2003, 4(2): 249-264 doi: 10.1093/biostatistics/4.2.249 [13] DE OLIVEIRA DAL'MOLIN C G, QUEK L E, PALFREYMAN R W, et al. AraGEM, a genome-scale reconstruction of the primary metabolic network in Arabidopsis[J]. Plant Physiology, 2010, 152(2): 579-589 doi: 10.1104/pp.109.148817 [14] SIRIWACH R, MATSUDA F, YANO K, et al. Drought stress responses in context-specific genome-scale metabolic models of Arabidopsis thaliana[J]. Metabolites, 2020, 10(4): 159 doi: 10.3390/metabo10040159 [15] EBRAHIM A, LERMAN J A, PALSSON B O, et al. COBRApy: constraints-based reconstruction and analysis for python[J]. BMC Systems Biology, 2013, 7(1): 74 doi: 10.1186/1752-0509-7-74 [16] BORDEL S. Constraint based modeling of metabolism allows finding metabolic cancer hallmarks and identifying personalized therapeutic windows[J]. Oncotarget, 2018, 9(28): 19716-19729 doi: 10.18632/oncotarget.24805 [17] RAŠKEVIČIUS V, MIKALAYEVA V, ANTANAVIČIŪTĖ I, et al. Genome scale metabolic models as tools for drug design and personalized medicine[J]. PLoS One, 2018, 13(1): e0190636 doi: 10.1371/journal.pone.0190636 [18] ORTH J D, THIELE I, PALSSON B Ø. What is flux balance analysis?[J]. Nature Biotechnology, 2010, 28(3): 245-248 doi: 10.1038/nbt.1614 [19] HUANG D W, SHERMAN B T, TAN Q N, et al. DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists[J]. Nucleic Acids Research, 2007, 35(S2): W169-W175 [20] SUN T H, ZHOU F, HUANG X Q, et al. ORANGE represses chloroplast biogenesis in etiolated Arabidopsis cotyledons via interaction with TCP14[J]. The Plant Cell, 2019, 31(12): 2996-3014 doi: 10.1105/tpc.18.00290 [21] VANDENBRINK J P, HERRANZ R, POEHLMAN W L, et al. RNA‐seq analyses of Arabidopsis thaliana seedlings after exposure to blue‐light phototropic stimuli in microgravity[J]. American Journal of Botany, 2019, 106(11): 1466-1476 doi: 10.1002/ajb2.1384 [22] PAUL A L, AMALFITANO C E, FERL R J. Plant growth strategies are remodeled by spaceflight[J]. BMC Plant Biology, 2012, 12(1): 232 doi: 10.1186/1471-2229-12-232 [23] AKRAM M. Citric acid cycle and role of its intermediates in metabolism[J]. Cell Biochemistry and Biophysics, 2014, 68(3): 475-478 doi: 10.1007/s12013-013-9750-1 [24] MUÑOZ-BERTOMEU J, CASCALES-MIÑANA B, ALAIZ M, et al. A critical role of plastidial glycolytic Glyceraldehyde-3-phosphate dehydrogenase in the control of plant metabolism and development[J]. Plant Signaling & Behavior, 2010, 5(1): 67-69 [25] MELESHKO G I, ANTON’YAN A A, SYCHEV V N, et al. The effect of space flight factors on the pigment systems of one-celled algae[J]. USSR Space Life Sci Digest, 1991, 31: 43-45 [26] ABILOV Z K, ALEKPEROV U K, MASHINSKIY A L, et al. The morphological and functional state of the photosynthetic system of plant cells grown for varying periods under space flight condition[J]. USSR Space Life Sci Digest, 1986, 8: 15-18 [27] KORDYUM E, ADAMCHUK N. Clinorotation affects the state of photosynthetic membranes in Arabidopsis thaliana (L.) Heynh[J]. Journal of Gravitational Physiology: A Journal of the International Society for Gravitational Physiology, 1997, 4(2): P77-P78 [28] TRIPATHY B C, BROWN C S, LEVINE H G, et al. Growth and photosynthetic responses of wheat plants grown in space[J]. Plant Physiology, 1996, 110(3): 801-806 doi: 10.1104/pp.110.3.801 [29] JIAO S X, HILAIRE E, PAULSEN A Q, et al. Brassica rapa plants adapted to microgravity with reduced photosystem I and its photochemical activity[J]. Physiologia Plantarum, 2004, 122(2): 281-290 doi: 10.1111/j.1399-3054.2004.00400.x -
-






 下载:
下载: