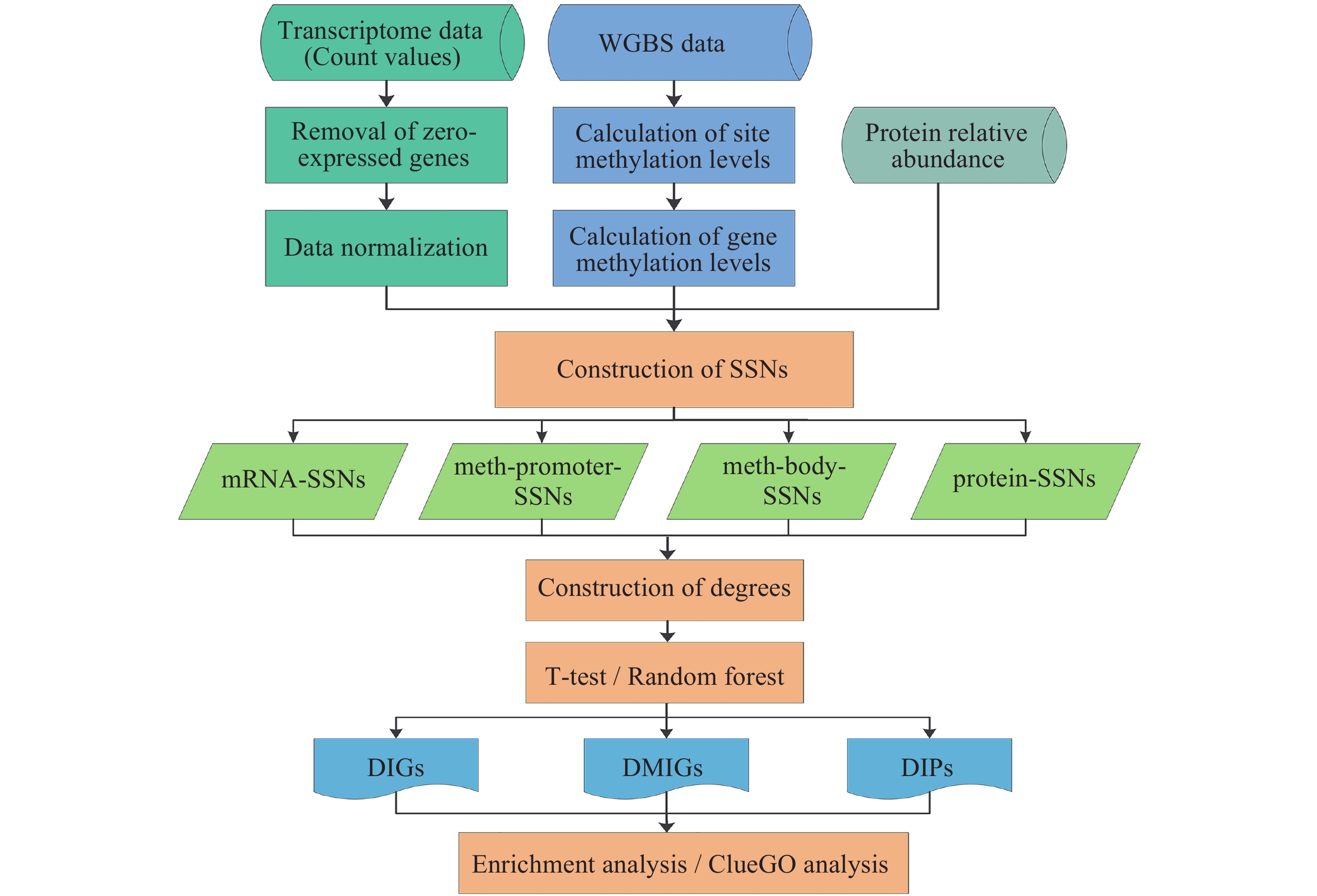
空间飞行条件下小鼠不同组织中多组学分子互作模式挖掘及关键基因识别
doi: 10.11728/cjss2025.02.2024-0137 cstr: 32142.14.cjss.2024-0137
Mining of Multi-omics Molecular Interaction Patterns and Identification of Key Genes in Multiple Mouse Tissues under Spaceflight Conditions
-
摘要: 为了从系统生物学的角度探究空间生物学效应, 本文基于单样本网络(Single-Sample Network, SSN)开发了一种生物信息学分析方法. 首先, 为来自空间飞行小鼠不同组织的每个样本分别构建了转录组、DNA甲基化、蛋白质组层面的SSN. 然后, 提取每个SSN中所有节点的拓扑特征, 并使用T检验识别出不同组学层面的差异互作分子. 结果表明, 虽然不同组学层面互作模式改变的分子交集有限, 但其调控的生物过程和通路具有相似性, 主要包括代谢过程、DNA损伤响应、细胞周期、氧化应激、昼夜节律等. 本文还分别构建了不同组学层面的共互作网络, 并识别了枢纽(Hub)基因. 此外, 空间飞行改变的分子互作模式可能与一系列疾病发生和病毒重激活有关.Abstract: To explore the space biological effects from a systems biology perspective, a bioinformatics analysis pipeline was developed based on Single-Sample Network (SSN) and was used to mine multi-omics molecular interaction patterns and key genes in multiple mouse tissues under spaceflight conditions. First, we collected four spaceflight mouse datasets from the GeneLab platform, which included transcriptome, DNA methylation, and proteome sequencing results from the adrenal gland, kidney, liver, and quadriceps. For each sample, four SSNs (mRNA-SSN, meth-promoter-SSN, meth-body-SSN, protein-SSN) were constructed. Next, the topological features of the nodes in each SSN were extracted, and a T-test was performed to identify the molecules with altered interaction patterns across different omics levels under spaceflight conditions. The results indicated that, although the overlap of molecules with altered interaction patterns across different omics levels was limited, the biological processes and pathways they regulated exhibited similarities, which primarily included metabolic processes, DNA damage response, cell cycle, oxidative stress, and circadian rhythms. Notably, the protein/amino acid metabolic process and nucleic acid (DNA/RNA) metabolic process appeared in multiple tissues and showed high significance. The key genes involved in multi-omics regulation were identified, including Park7, Tmed3, Rbbp7, Hunk, Rad23a, Cd36, etc. Furthermore, we constructed co-interaction networks for the transcriptome and DNA methylation, transcriptome and proteome, as well as for all three omics layers, and identified the Hub genes (such as Rbbp7, Egfr, Rpl4, Srms, Cabp4) from them. Functional analysis revealed that several key/Hub genes were involved in regulating the aforementioned biological processes. To gain further insight into the expression patterns of key/Hub genes, additional datasets from the same tissue in the GeneLab database were analyzed using DESeq2. The results revealed that several genes were also differentially expressed under spaceflight conditions in other datasets. The molecular interaction patterns altered by spaceflight might be associated with the occurrence of various diseases (such as cancer, neurological disorders, neurodegenerative diseases, cardiovascular diseases, and diabetes) and the reactivation of viruses (such as Epstein-Barr virus, herpes simplex virus, and human cytomegalovirus).
-
Key words:
- Space environment /
- Single-sample network /
- Molecular interaction pattern /
- Multi-omics
-
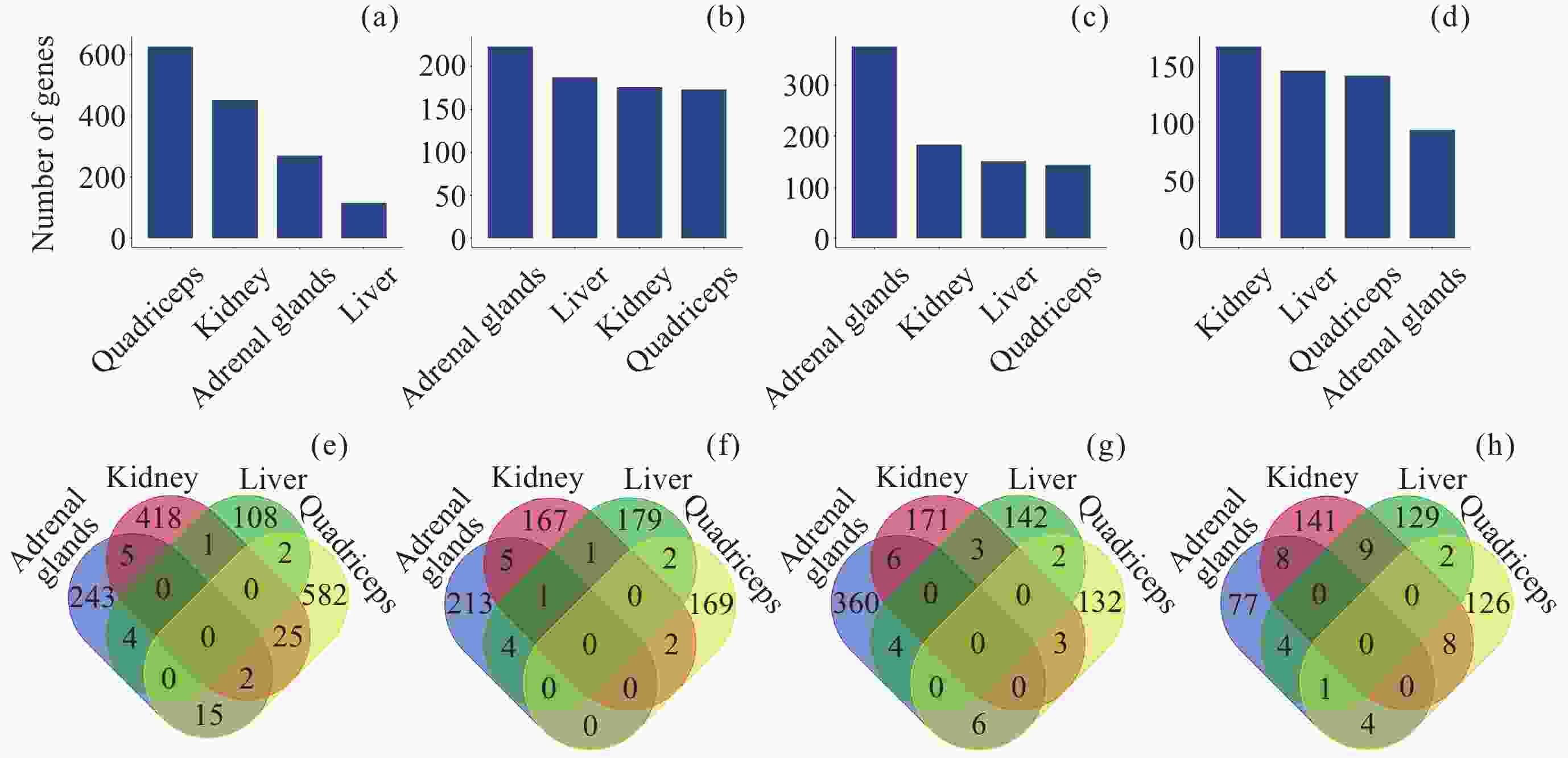
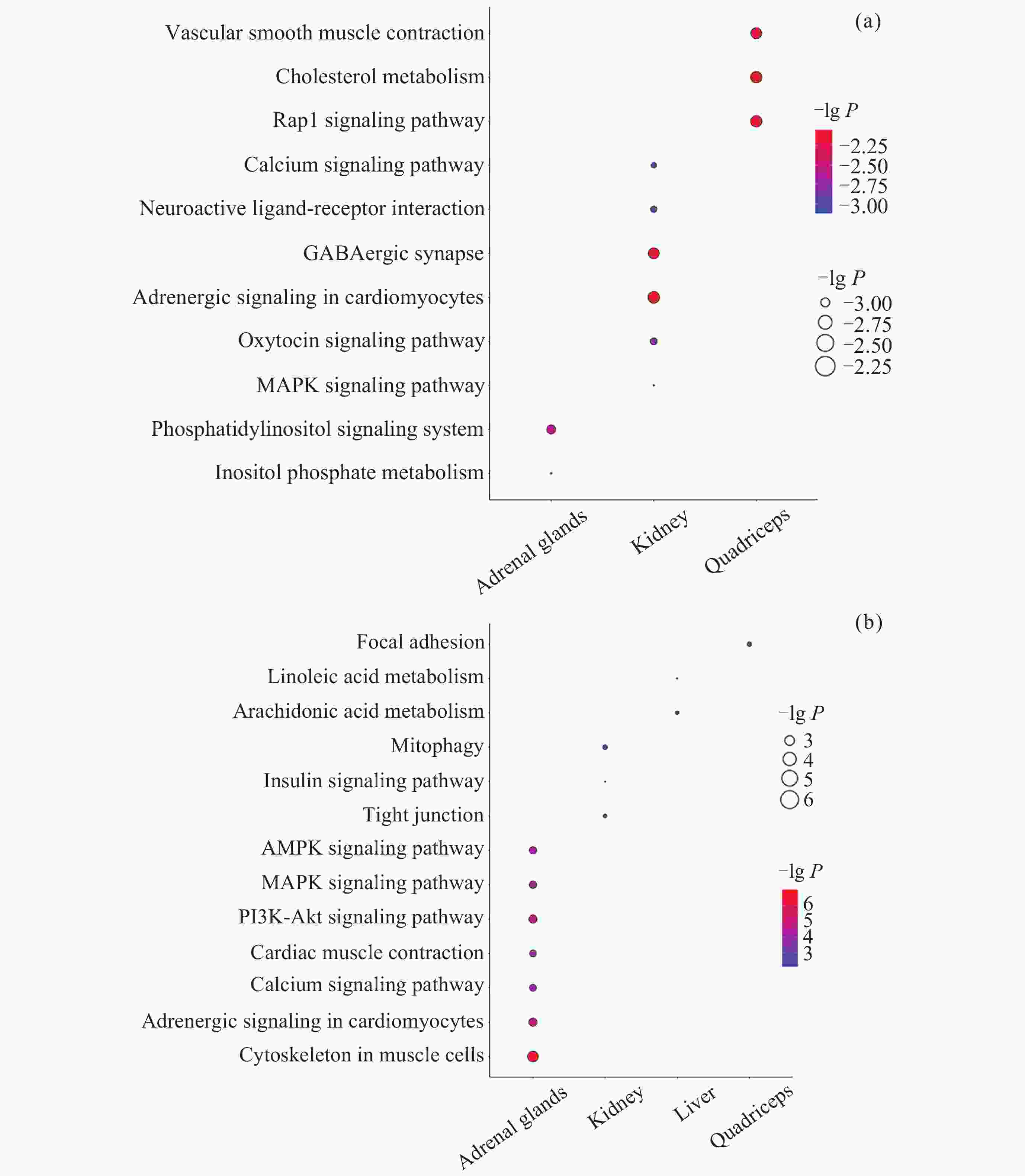
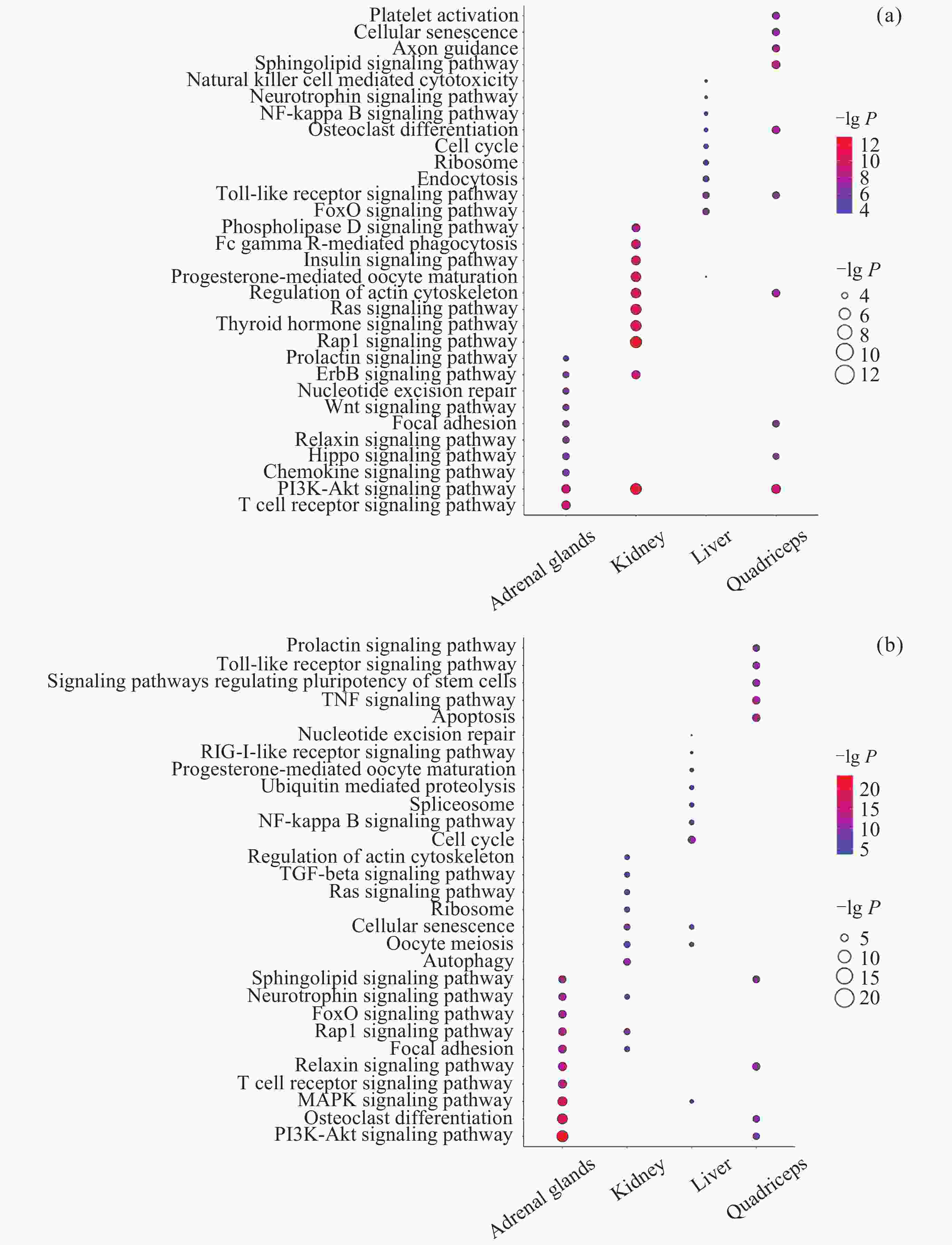
图 2 差异互作分子的数量及交集. (a) DIG数量, (b) 启动子中DMIG数量, (c) 基因区中DMIG数量, (d) DIP数量, (e)~(h) 转录组、甲基化(启动子)、甲基化(基因体)和蛋白质组中不同组织差异互作分子的交集
Figure 2. Number and intersection of differentially interacted molecules. (a) Number of DIGs, (b) number of DMIGs in promoters, (c) number of DMIGs in gene bodies, (d) number of DIPs, (e)~(h) intersection of differentially interacted molecules among transcriptome, methylation (promoter), methylation (gene body), and proteome in various tissues
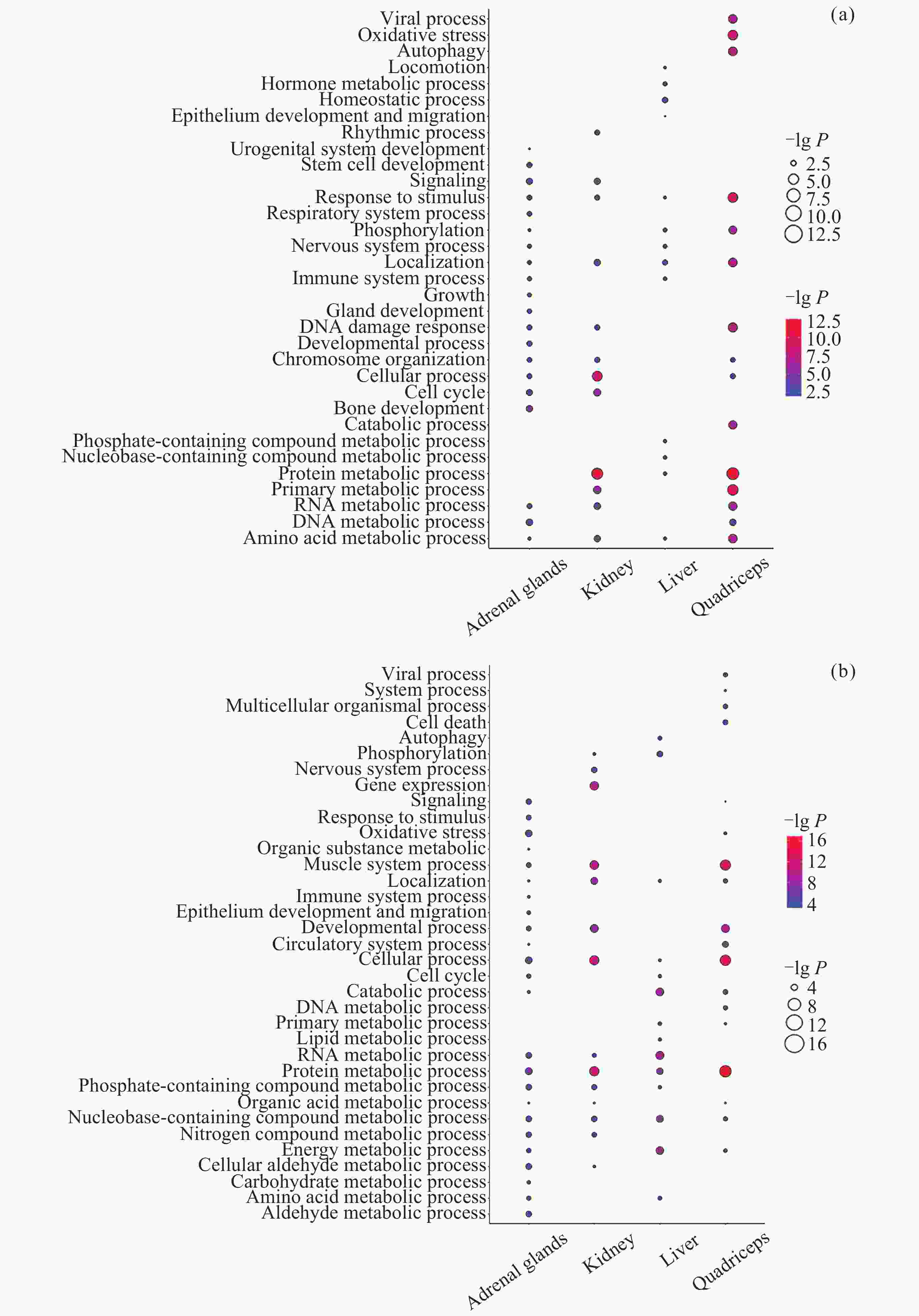
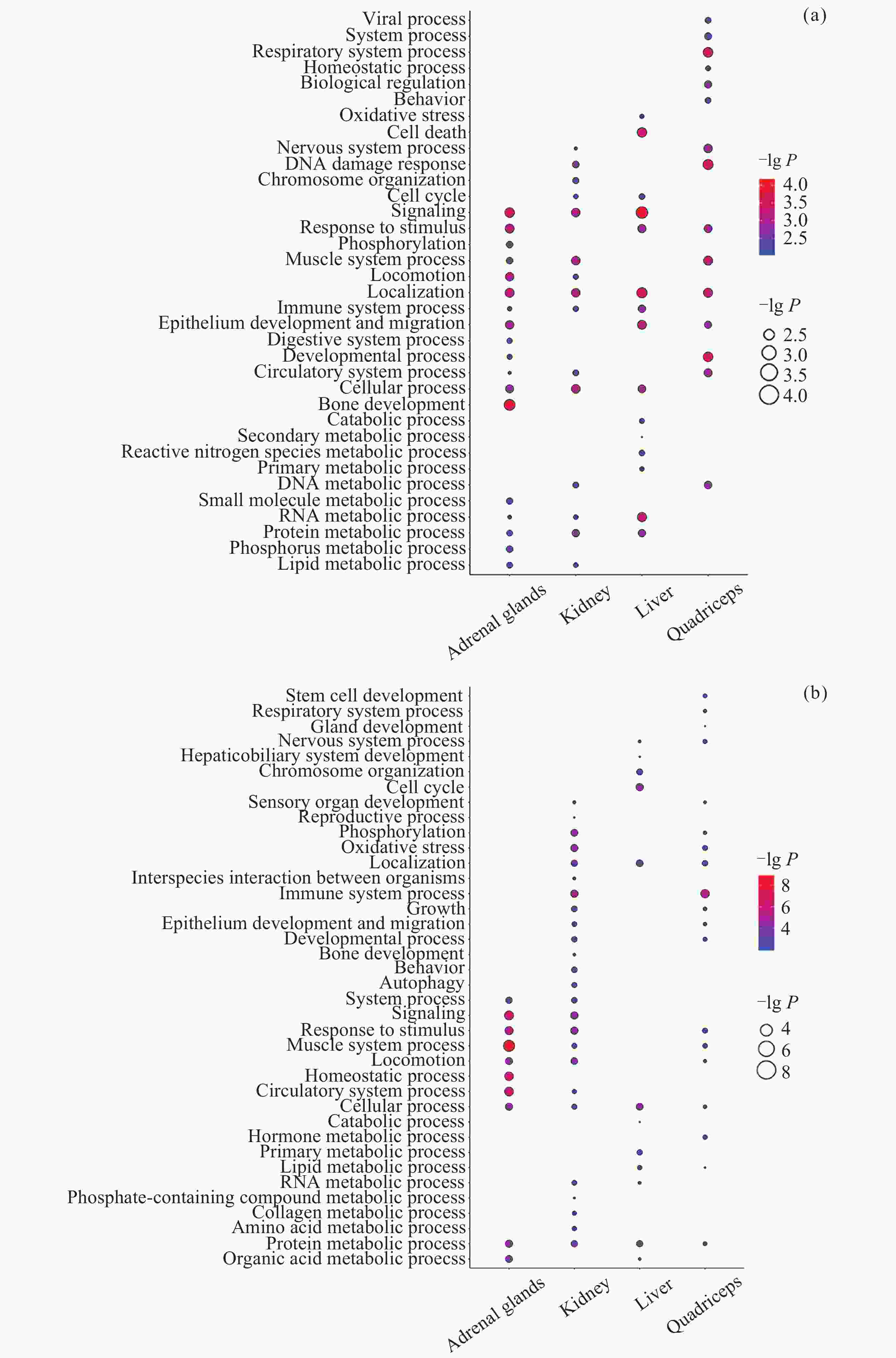
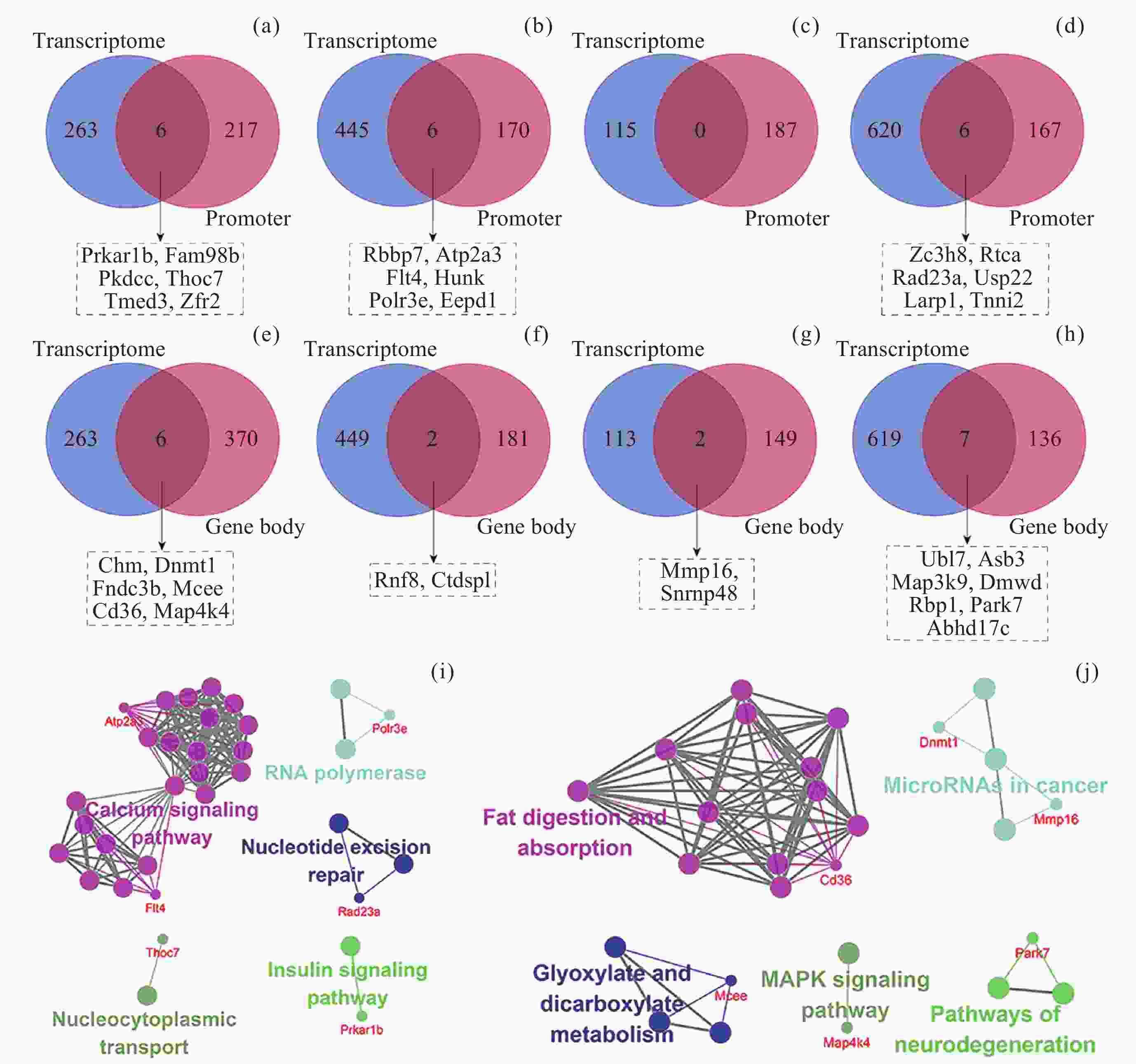
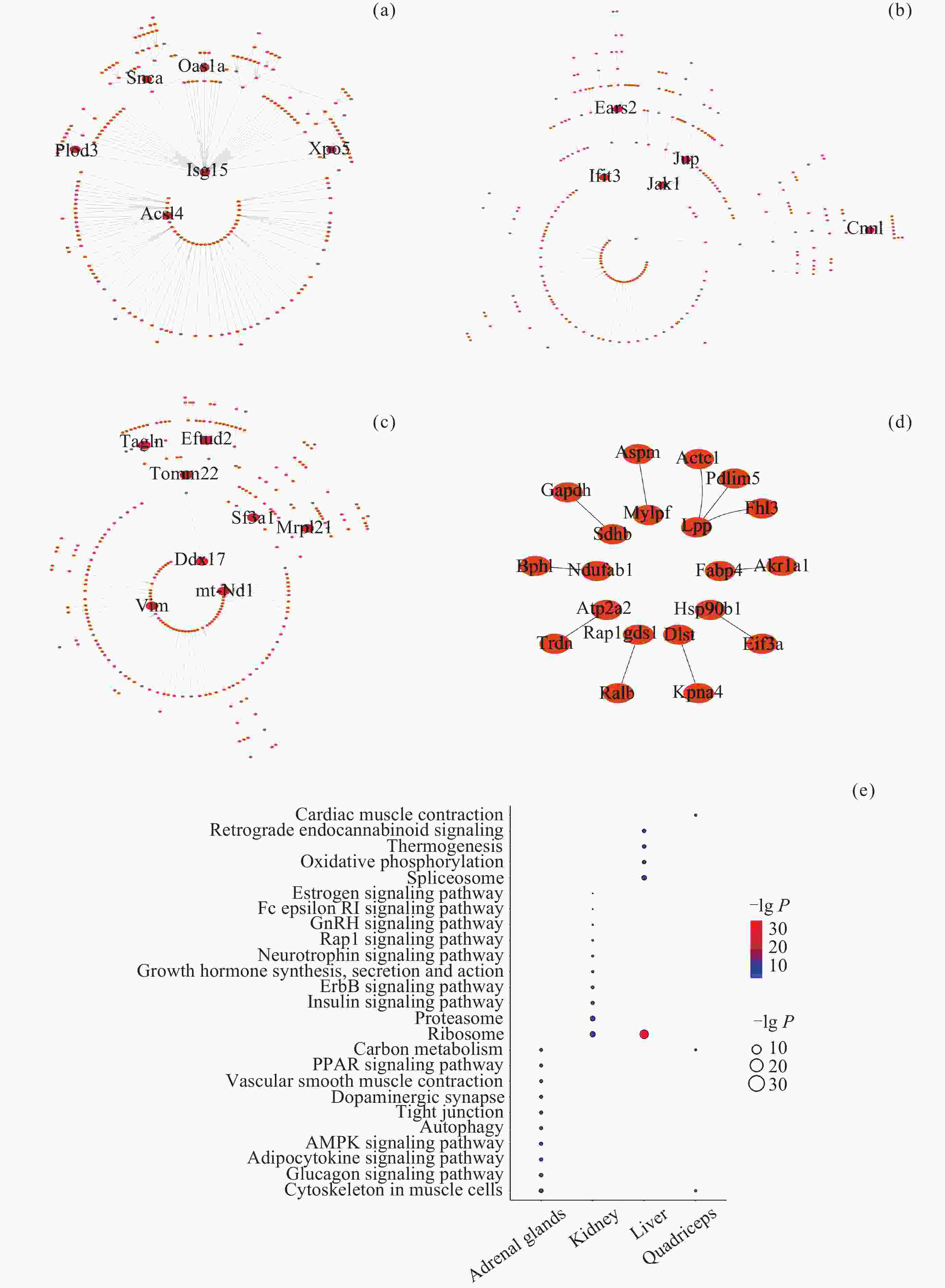
图 7 不同组织中DIG与DMIG的交集. (a)~(d)为DIG与启动子中DMIG的韦恩图, (e)~(h)为DIG与基因区中DMIG的韦恩图. (a)(e) 肾上腺, (b)(f) 肾, (c)(g) 肝, (d)(h) 股四头肌. (i) DIG与启动子中DMIG的交集基因参与的通路. (j) DIG与基因区中DMIG的交集基因参与的通路. 参与通路的基因以红色字体表示, 仅显示了ClueGO标注的overview pathway的名称
Figure 7. Intersection of DIGs and DMIGs in different tissues. (a)~(d) Venn diagrams of DIGs and DMIGs in the promoters, (e)~(h) Venn diagrams of DIGs and DMIGs in the gene bodies. (a)(e) Adrenal glands, (b)(f) Kidney, (c)(g) Liver, (d)(h) Quadriceps. (i) The pathways involving genes in the intersection of DIGs and DMIGs in the promoters, (j) the pathways involving genes in the intersection of DIGs and DMIGs in the gene bodies. Genes participating in the pathway were indicated in red font. Only the names of overview pathways selected by ClueGO were annotated
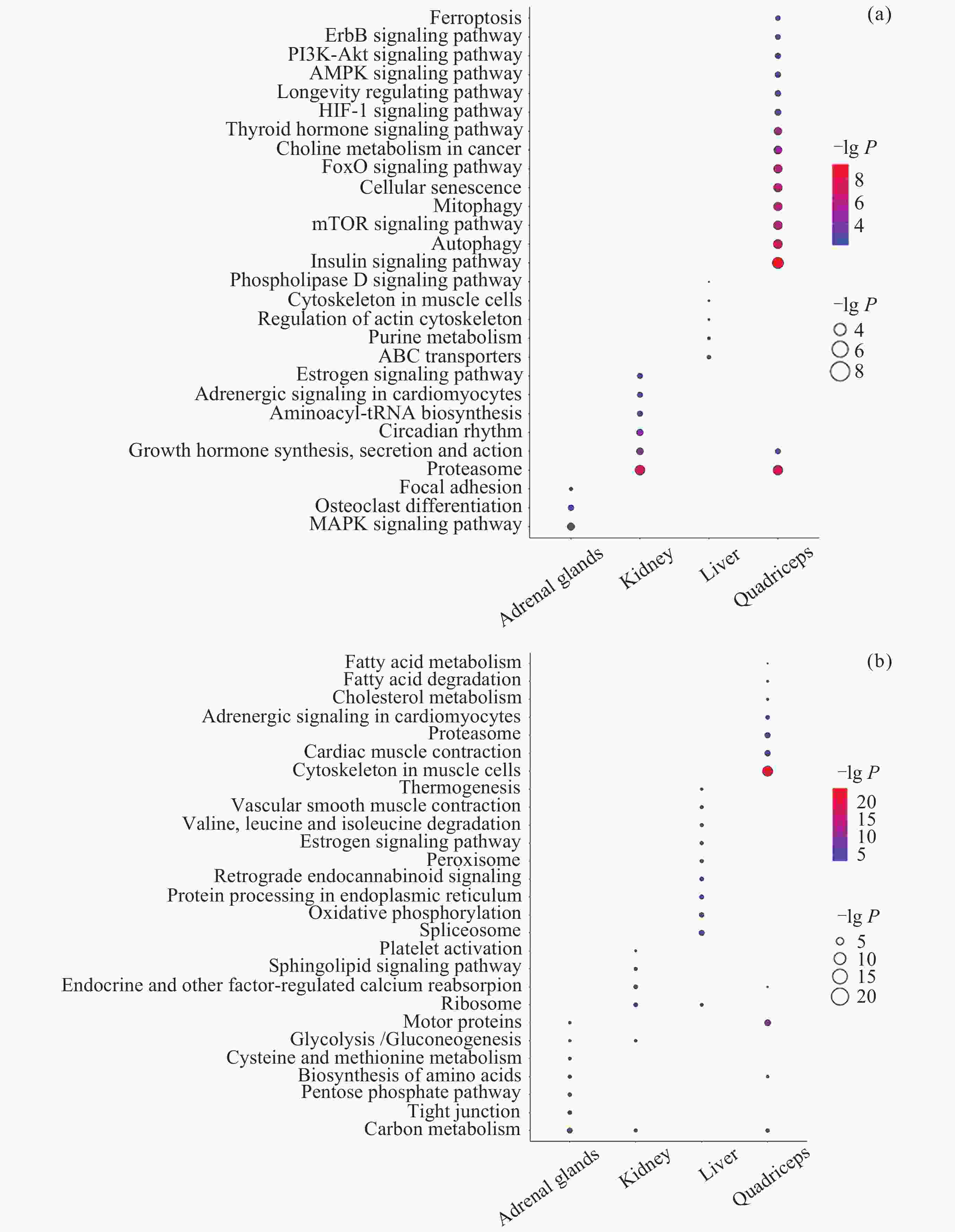
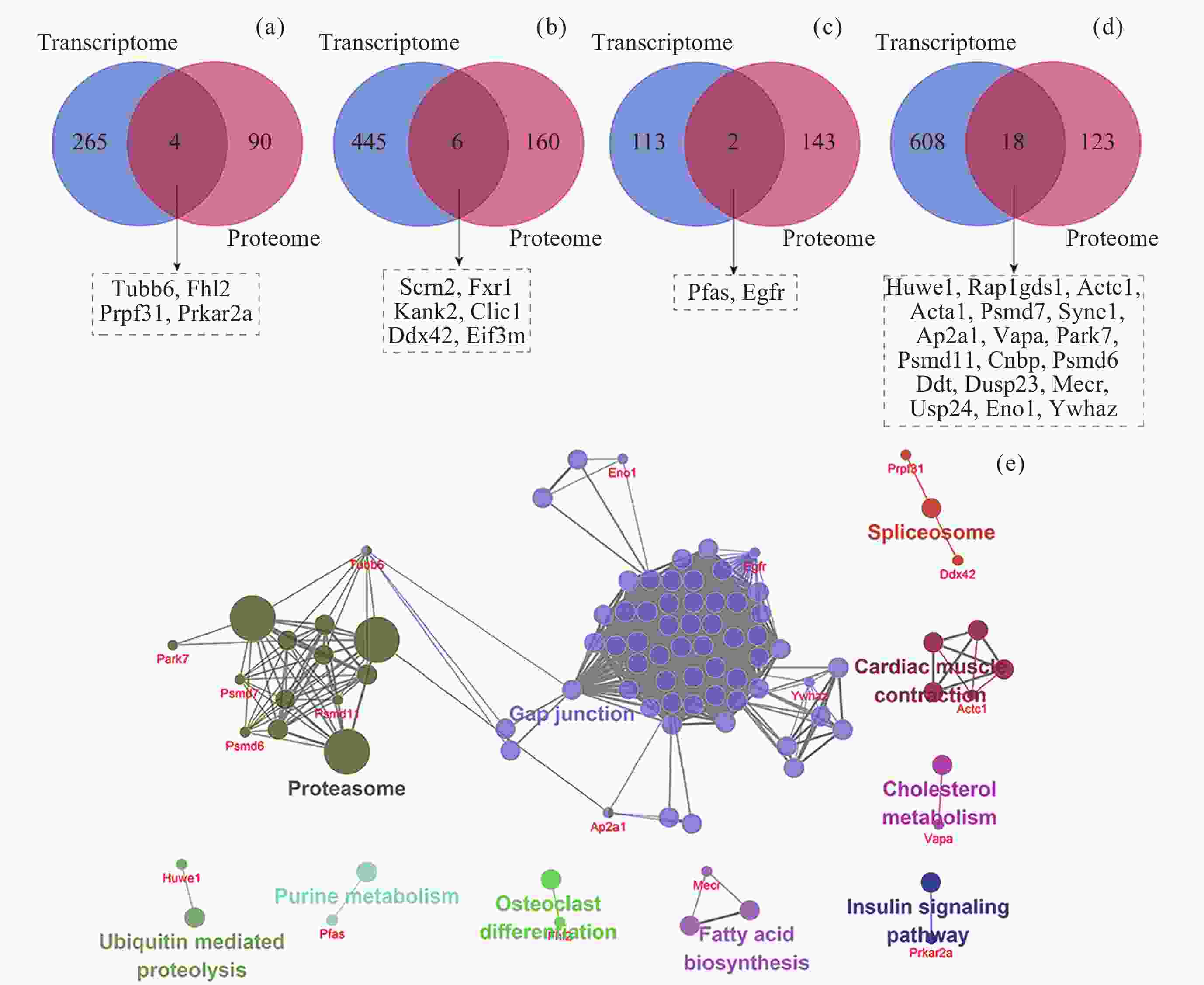
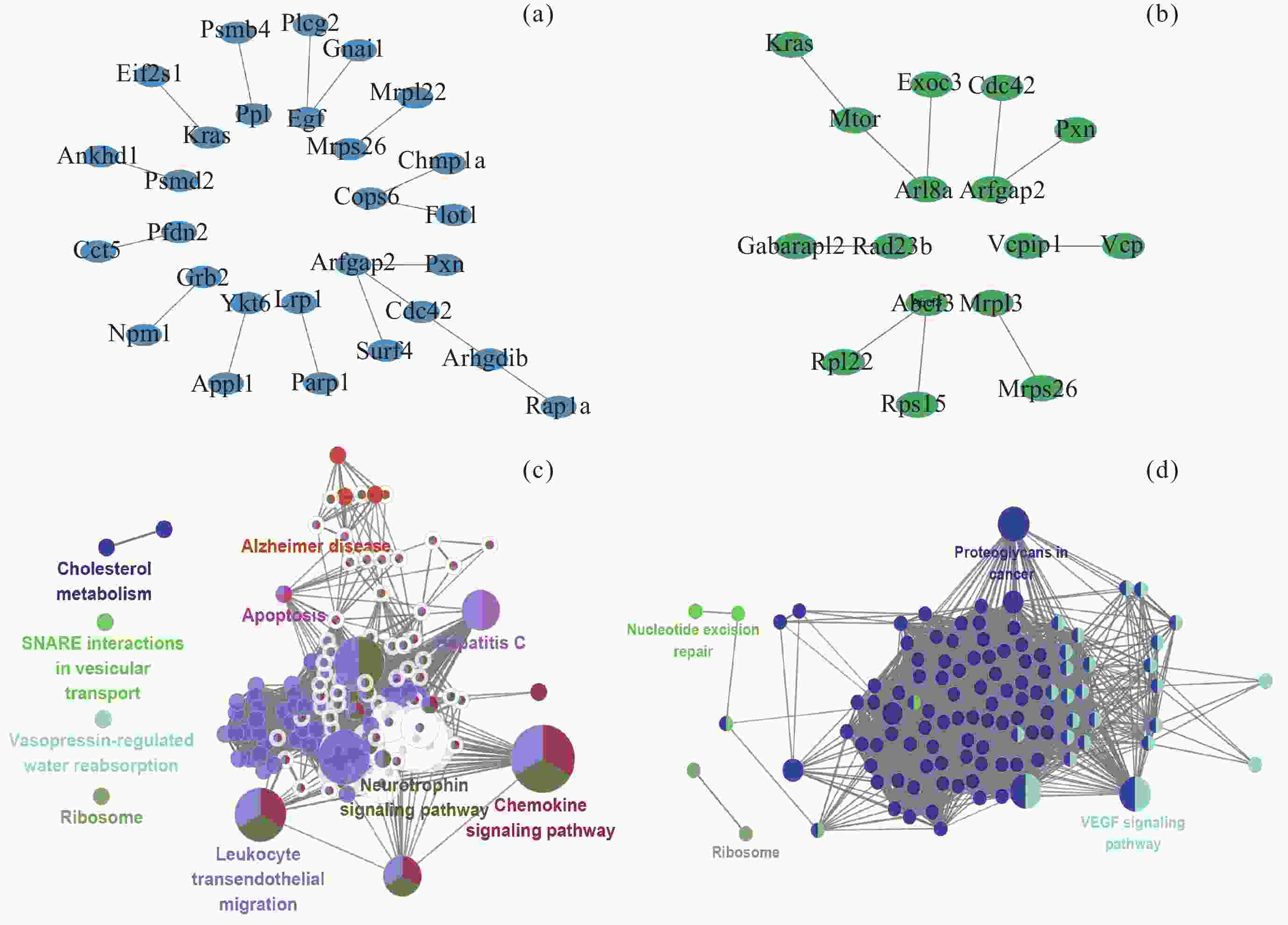
图 11 转录组和蛋白质组层面的共互作网络. (a) 肾上腺, (b) 肾, (c) 肝, (d) 股四头肌, (e) 上述4个共互作网络参与的通路. 红色节点代表Hub基因
Figure 11. Co-interaction networks at the transcriptome and proteome levels. (a) Adrenal glands, (b) kidney, (c) liver, (d) quadriceps, (e) the pathways of above four co-interaction networks. The red nodes represent Hub genes
图 12 转录组、甲基化和蛋白质组层面的共互作网络. (a) 肾脏中转录组、甲基化(启动子)和蛋白质组层面的共互作网络, (b) 肾脏中转录组、甲基化(基因区)和蛋白质组层面的共互作网络, (c) A网络参与的通路, (d) B网络参与的通路
Figure 12. Co-interaction networks at the transcriptome, methylation and proteome levels. (a) Co-interaction networks at the transcriptome, methylation (promoter) and proteome levels in the kidneys, (b) co-interaction networks at the transcriptome, methylation (gene body) and proteome levels in the kidneys, (c) pathways of network A, (d) pathways of network B
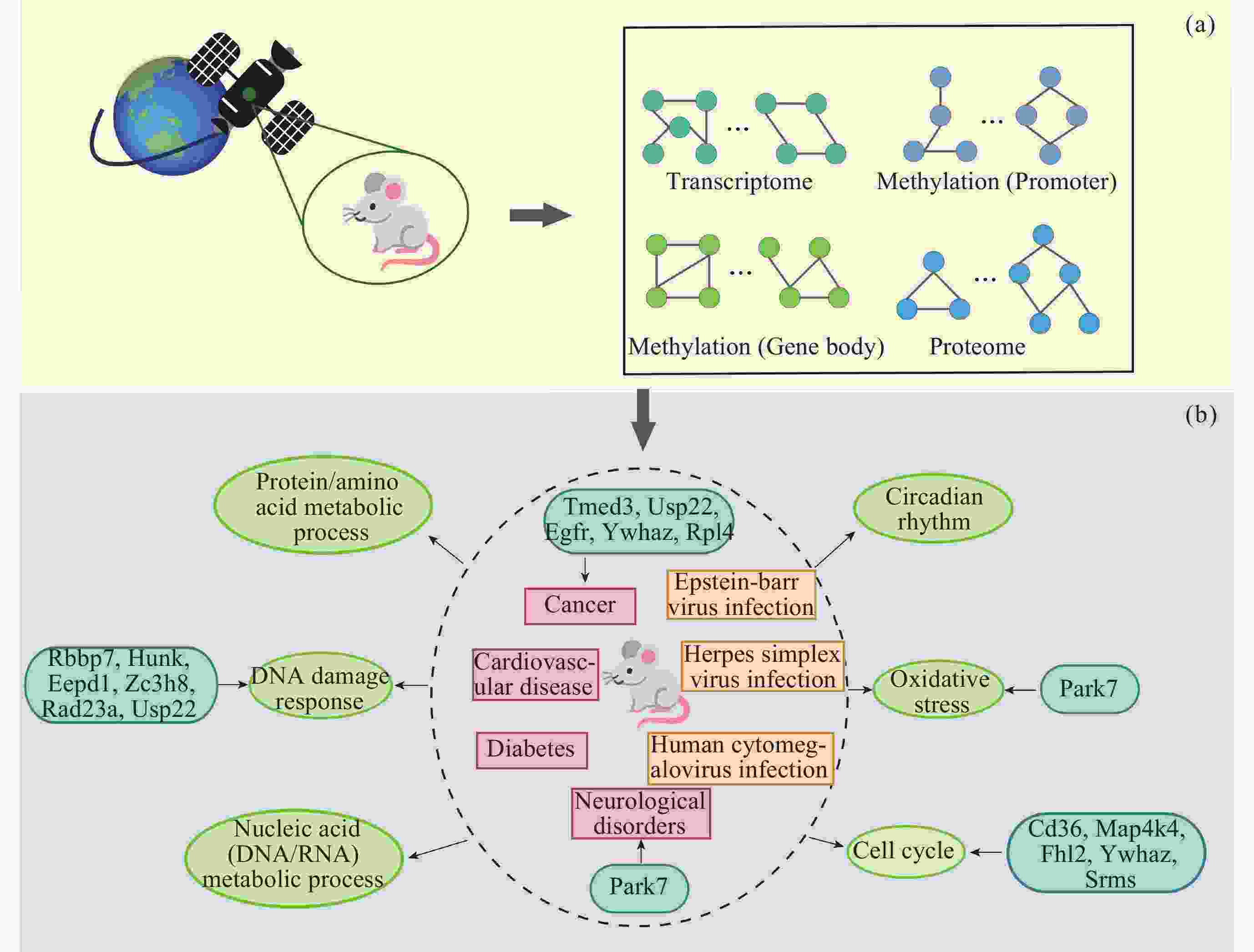
图 13 基于SSN的生物信息学分析流程及主要结果. 浅绿色圆圈代表生物过程/通路, 红色方框代表疾病, 黄色方框代表病毒感染, 深绿色圆角方框代表参与该过程的关键/Hub基因
Figure 13. Bioinformatics analysis workflow based on SSN and its main results. The light green circles represent biological processes/pathways, the red squares represent diseases, the yellow squares represent viral infections, and the dark green rounded squares represent the key/Hub genes involved in these processes
表 1 本文使用的GeneLab数据集的样本个数
Table 1. Number of samples in the GeneLab dataset used in this article
数据集编号 组织 转录组 (GC/SF) 甲基化 (GC/SF) 蛋白质组 (GC/SF) OSD-47 肝 3/3 3/3 – OSD-98 肾上腺 5/6 5/6 4/4 OSD-102 肾 6/6 6/6 6/6 OSD-103 股四头肌 6/6 6/6 6/6 OSD-137 肝 6/6 – 6/6 表 2 疾病/病毒相关的KEGG通路
Table 2. Disease/virus-related KEGG pathways
通路 P 组织 组学 单纯疱疹病毒 1 型感染 $ 2.21\times {10}^{-4} $ 肾上腺 转录组 朊病毒病 $ 3.40\times {10}^{-7} $ 肾 转录组 帕金森病 $ 3.61\times {10}^{-7} $ 肾 转录组 肌萎缩侧索硬化症 $ 4.76\times {10}^{-6} $ 肾 转录组 神经退行性病变的途径 $ 1.12\times {10}^{-5} $ 肾 转录组 阿尔茨海默病 $ 2.68\times {10}^{-5} $ 肾 转录组 亨廷顿病 $ 2.93\times {10}^{-5} $ 肾 转录组 脊髓小脑共济失调 $ 4.23\times {10}^{-5} $ 肾 转录组 爱泼斯坦–巴尔病毒感染 $ 2.2\times {10}^{-9} $ 股四头肌 转录组 脊髓小脑共济失调 $ 3.85\times {10}^{-9} $ 股四头肌 转录组 亨廷顿病 $ 8.24\times {10}^{-8} $ 股四头肌 转录组 癌症中的通路 $ 5.42\times {10}^{-7} $ 股四头肌 转录组 麻疹 $ 9.20\times {10}^{-7} $ 股四头肌 转录组 神经退行性病变的途径 $ 1.41\times {10}^{-6} $ 股四头肌 转录组 肌萎缩侧索硬化症 $ 4.27\times {10}^{-6} $ 股四头肌 转录组 急性髓系白血病 $ 4.54\times {10}^{-6} $ 股四头肌 转录组 阿尔茨海默病 $ 8.52\times {10}^{-6} $ 股四头肌 转录组 慢性髓系白血病 $ 1.03\times {10}^{-5} $ 股四头肌 转录组 乙型肝炎 $ 2.06\times {10}^{-5} $ 股四头肌 转录组 丙型肝炎 $ 2.58\times {10}^{-5} $ 股四头肌 转录组 化学致癌作用 $ 1.08\times {10}^{-4} $ 股四头肌 转录组 前列腺癌 $ 1.37\times {10}^{-4} $ 股四头肌 转录组 甲型流感 $ 1.52\times {10}^{-4} $ 股四头肌 转录组 胰腺癌 $ 3.15\times {10}^{-4} $ 股四头肌 转录组 朊病毒病 $ 3.56\times {10}^{-4} $ 股四头肌 转录组 帕金森病 $ 3.73\times {10}^{-4} $ 股四头肌 转录组 胃癌 $ 3.90\times {10}^{-4} $ 股四头肌 转录组 人类巨细胞病毒感染 $ 4.07\times {10}^{-4} $ 股四头肌 转录组 耶尔森菌感染 $ 5.49\times {10}^{-4} $ 股四头肌 转录组 人乳头瘤病毒感染 $ 7.28\times {10}^{-4} $ 股四头肌 转录组 卡波西肉瘤相关疱疹病毒感染 $ 7.33\times {10}^{-4} $ 股四头肌 转录组 病毒致癌作用 $ 9.29\times {10}^{-4} $ 股四头肌 转录组 非小细胞肺癌 $ 1.03\times {10}^{-3} $ 股四头肌 转录组 肌萎缩侧索硬化症 $ 7.87\times {10}^{-7} $ 肾上腺 蛋白质组 沙门氏菌感染 $ 1.25\times {10}^{-4} $ 肾上腺 蛋白质组 神经退化途径 $ 2.69\times {10}^{-4} $ 肾上腺 蛋白质组 亨廷顿病 $ 4.23\times {10}^{-4} $ 肾上腺 蛋白质组 神经退化途径 $ 5.53\times {10}^{-6} $ 肾 蛋白质组 亨廷顿病 $ 3.03\times {10}^{-5} $ 肾 蛋白质组 肌萎缩侧索硬化症 $ 3.86\times {10}^{-5} $ 肾 蛋白质组 沙门氏菌感染 $ 1.62\times {10}^{-4} $ 肾 蛋白质组 肌萎缩侧索硬化症 $ 1.98\times {10}^{-7} $ 股四头肌 蛋白质组 朊病毒病 $ 2.74\times {10}^{-7} $ 股四头肌 蛋白质组 帕金森病 $ 2.85\times {10}^{-7} $ 股四头肌 蛋白质组 神经退化途径 $ 7.08\times {10}^{-7} $ 股四头肌 蛋白质组 亨廷顿病 $ 9.85\times {10}^{-7} $ 股四头肌 蛋白质组 脊髓小脑共济失调 $ 4.45\times {10}^{-6} $ 股四头肌 蛋白质组 阿尔茨海默病 $ 1.08\times {10}^{-4} $ 股四头肌 蛋白质组 沙门氏菌感染 $ 2.75\times {10}^{-4} $ 股四头肌 蛋白质组 肥厚性心肌病 $ 5.30\times {10}^{-4} $ 股四头肌 蛋白质组 扩张型心肌病 $ 6.07\times {10}^{-4} $ 股四头肌 蛋白质组 神经退化途径 $ 9.85\times {10}^{-11} $ 肝 蛋白质组 肌萎缩侧索硬化症 $ 4.19\times {10}^{-8} $ 肝 蛋白质组 化学致癌作用 $ 5.54\times {10}^{-8} $ 肝 蛋白质组 帕金森病 $ 3.86\times {10}^{-7} $ 肝 蛋白质组 阿尔茨海默病 $ 4.37\times {10}^{-7} $ 肝 蛋白质组 非酒精性脂肪肝病 $ 1.52\times {10}^{-5} $ 肝 蛋白质组 亨廷顿病 $ 4.96\times {10}^{-5} $ 肝 蛋白质组 朊病毒病 $ 1.06\times {10}^{-4} $ 肝 蛋白质组 糖尿病性心肌病 $ 1.20\times {10}^{-4} $ 肝 蛋白质组 亨廷顿病 $ 1.09\times {10}^{-3} $ 肾上腺 启动子 肌萎缩侧索硬化症 $ 4.54\times {10}^{-3} $ 肾上腺 启动子 帕金森病 $ 6.37\times {10}^{-3} $ 肾上腺 启动子 神经退化途径 $ 9.12\times {10}^{-3} $ 肾上腺 启动子 致心律失常性右心室心肌病 $ 5.26\times {10}^{-3} $ 肾 启动子 肥厚性心肌病 $ 8.90\times {10}^{-3} $ 肾 启动子 扩张型心肌病 $ 9.85\times {10}^{-3} $ 肾 启动子 尼古丁成瘾 $ 3.02\times {10}^{-4} $ 肾 启动子 吗啡成瘾 $ 9.14\times {10}^{-4} $ 肾 启动子 范康尼贫血途径 $ 8.62\times {10}^{-3} $ 肾 启动子 甲状腺癌 $ 3.70\times {10}^{-3} $ 肝 启动子 查加斯病 $ 1.80\times {10}^{-3} $ 肝 启动子 肥厚性心肌病 $ 8.46\times {10}^{-6} $ 肾上腺 基因区 扩张型心肌病 $ 1.10\times {10}^{-5} $ 肾上腺 基因区 人乳头瘤病毒感染 $ 1.68\times {10}^{-4} $ 肾上腺 基因区 糖尿病性心肌病 $ 4.84\times {10}^{-4} $ 肾 基因区 胰岛素抵抗 $ 2.29\times {10}^{-3} $ 肾 基因区 II 型糖尿病 $ 7.46\times {10}^{-3} $ 肾 基因区 人类免疫缺陷病毒 1 型感染 $ 4.34\times {10}^{-3} $ 股四头肌 基因区 肾细胞癌 $ 9.91\times {10}^{-3} $ 股四头肌 基因区 阿米巴病 $ 6.18\times {10}^{-4} $ 股四头肌 基因区 人类 T 细胞白血病病毒 1 型感染 $ 4.88\times {10}^{-3} $ 股四头肌 基因区 表 3 关键基因的表达情况
Table 3. Expression of key genes
基因 组织 P-value 数据集 Rbbp7 肾 0.0228 OSD-163 Polr3e 肾 0.0081 OSD-163 Ctdspl 肾 0.0164 OSD-163 Eif3 m 肾 0.0008 OSD-163 Kank2 肾 0.0444 OSD-163 Hunk 肾 0.0458 OSD-253 Snrnp48 肝 0.0064 OSD-168 Egfr 肝 0.0237 OSD-168 Egfr 肝 0.0072 OSD-173 Rad23a 肌肉 0.0003 OSD-99 Asb3 肌肉 0.0003 OSD-99 Map3k9 肌肉 0.0001 OSD-99 Park7 肌肉 0.0005 OSD-99 Abhd17c 肌肉 0.03597 OSD-99 Acta1 肌肉 0.0315 OSD-99 Psmd7 肌肉 0.0357 OSD-99 Syne1 肌肉 0.0060 OSD-99 Vapa 肌肉 0.0413 OSD-99 Psmd11 肌肉 0.0033 OSD-99 Cnbp 肌肉 $ 4.80\times {10}^{-5} $ OSD-99 Ddt 肌肉 0.0206 OSD-99 Dusp23 肌肉 $ 1.29\times {10}^{-5} $ OSD-99 Mecr 肌肉 $ 3.43\times {10}^{-6} $ OSD-99 Eno1 肌肉 $ 3.70\times {10}^{-5} $ OSD-99 Ywhaz 肌肉 $ 1.25\times {10}^{-8} $ OSD-99 Usp22 肌肉 0.0422 OSD-101 Dmwd 肌肉 0.0129 OSD-101 Cnbp 肌肉 0.0236 OSD-101 Ywhaz 肌肉 0.0014 OSD-101 Zc3h8 肌肉 0.0231 OSD-104 Tnni2 肌肉 $ 1.90\times {10}^{-13} $ OSD-104 Dmwd 肌肉 $ 2.03\times {10}^{-14} $ OSD-104 Park7 肌肉 0.0041 OSD-104 Rap1gds1 肌肉 0.0295 OSD-104 Acta1 肌肉 0.0326 OSD-104 Psmd7 肌肉 $ 9.03\times {10}^{-22} $ OSD-104 Psmd11 肌肉 $ 1.61\times {10}^{-5} $ OSD-104 Cnbp 肌肉 $ 1.11\times {10}^{-6} $ OSD-104 Ddt 肌肉 0.0002 OSD-104 Dusp23 肌肉 0.0017 OSD-104 Mecr 肌肉 0.0216 OSD-104 Usp24 肌肉 $ 1.85\times {10}^{-5} $ OSD-104 Eno1 肌肉 $ 3.01\times {10}^{-9} $ OSD-104 Ywhaz 肌肉 0.0166 OSD-104 Usp22 肌肉 0.0056 OSD-105 Larp1 肌肉 0.0276 OSD-105 Map3k9 肌肉 0.0364 OSD-105 Syne1 肌肉 0.0088 OSD-105 Cnbp 肌肉 $ 9.57\times {10}^{-6} $ OSD-105 Ddt 肌肉 0.0011 OSD-105 Dusp23 肌肉 0.0020 OSD-105 Eno1 肌肉 $ 5.66\times {10}^{-5} $ OSD-105 Ywhaz 肌肉 $ 9.27\times {10}^{-5} $ OSD-105 Acta1 肌肉 0.0063 OSD-401 Syne1 肌肉 $ 4.63\times {10}^{-5} $ OSD-401 Cnbp 肌肉 0.0039 OSD-401 Ddt 肌肉 0.0135 OSD-401 Usp24 肌肉 0.0456 OSD-401 表 4 转录组和甲基化的共互作网络中的Hub基因
Table 4. Hub genes in the co-interaction networks at the transcriptome and methylation levels
基因 度 组织 类型 Chd1 370 肾上腺 启动子 Rbbp7 110 肾上腺 启动子 Pparg 91 肾上腺 启动子 Kras 89 肾上腺 启动子 Egfr 87 肾上腺 启动子 Smarca4 87 肾上腺 启动子 Cul4a 99 肾 启动子 Nedd8 87 肾 启动子 Rpl8 82 肾 启动子 Rbbp7 77 肾 启动子 Rpl4 77 肾 启动子 Ranbp2 82 肝 启动子 Prkdc 78 肝 启动子 Srms 69 肝 启动子 Mcph1 66 肝 启动子 Crebbp 58 肝 启动子 Rpl10 58 肝 启动子 Tnf 156 股四头肌 启动子 Cabp4 82 股四头肌 启动子 Syf2 58 股四头肌 启动子 Ankrd10 51 股四头肌 启动子 Pik3r1 50 股四头肌 启动子 Ywhaz 129 肾上腺 基因区 Ppp2ca 113 肾上腺 基因区 Egfr 113 肾上腺 基因区 Ywhah 102 肾上腺 基因区 Myc 94 肾上腺 基因区 Ubb 107 肾 基因区 Rpl4 73 肾 基因区 Ppp1ca 65 肾 基因区 Baz1b 59 肾 基因区 Vcp 52 肾 基因区 Srms 70 肝 基因区 Top2b 30 肝 基因区 Plk4 28 肝 基因区 Mib2 24 肝 基因区 Ube2a 22 肝 基因区 Bgn 40 股四头肌 基因区 Ddit3 39 股四头肌 基因区 Ripk3 39 股四头肌 基因区 Cabp4 38 股四头肌 基因区 Nras 32 股四头肌 基因区 表 5 Hub基因的表达情况
Table 5. Expression of Hub genes
基因 组织 P-value 数据集 Rbbp7 肾 0.0228 OSD-163 Rpl8 肾 0.0038 OSD-163 Ppp1ca 肾 0.0045 OSD-163 Baz1b 肾 0.0168 OSD-163 Jup 肾 0.0357 OSD-163 Rpl4 肾 0.0400 OSD-163 Jup 肾 0.0082 OSD-163 Ifit3 肾 0.0172 OSD-163 Ears2 肾 0.0290 OSD-163 Cnn1 肾 0.0017 OSD-163 Ranbp2 肝 0.0177 OSD-168 Prkdc 肝 $ 3.14\times {10}^{-5} $ OSD-168 Mcph1 肝 0.0013 OSD-168 Rpl10 肝 $ 3.67\times {10}^{-6} $ OSD-168 Tomm22 肝 0.0029 OSD-168 Ddx17 肝 0.0031 OSD-168 Srms 肝 0.0132 OSD-168 Ranbp2 肝 0.0037 OSD-173 Mib2 肝 0.0393 OSD-173 Ddx17 肝 0.0115 OSD-173 Tagln 肝 0.0125 OSD-242 Crebbp 肝 0.0088 OSD-245 Srms 肝 0.0400 OSD-245 Tomm22 肝 0.0052 OSD-245 Vim 肝 0.0268 OSD-245 Srms 肝 0.0038 OSD-245 Mcph1 肝 0.0476 OSD-245 Crebbp 肝 $ 8.00\times {10}^{-5} $ OSD-245 Rpl10 肝 0.0024 OSD-245 Srms 肝 0.0038 OSD-245 Tomm22 肝 0.0029 OSD-245 Syf2 肌肉 $ 9.29\times {10}^{-5} $ OSD-99 Ankrd10 肌肉 0.0004 OSD-99 Bgn 肌肉 $ 9.13\times {10}^{-11} $ OSD-99 Ddit3 肌肉 0.0027 OSD-99 Ripk3 肌肉 0.0087 OSD-99 Tnf 肌肉 0.0324 OSD-101 Syf2 肌肉 0.0214 OSD-101 Ankrd10 肌肉 0.0352 OSD-101 Tnf 肌肉 0.0069 OSD-104 Syf2 肌肉 0.0026 OSD-104 Ankrd10 肌肉 0.0009 OSD-104 Pik3 r1 肌肉 0.0032 OSD-104 Bgn 肌肉 $ 3.30\times {10}^{-5} $ OSD-104 Ddit3 肌肉 0.0196 OSD-104 Ripk3 肌肉 0.0268 OSD-104 Syf2 肌肉 0.0491 OSD-105 Nras 肌肉 0.0320 OSD-105 Bgn 肌肉 0.0272 OSD-401 -
[1] 轩莹莹, 杨玉田, 孙月红, 等. 白藜芦醇对后肢去负荷雄性大鼠生殖损伤的对抗作用[J]. 空间科学学报, 2024, 44(1): 133-141 doi: 10.11728/cjss2024.01.2023-0063XUAN Yingying, YANG Yutian, SUN Yuehong, et al. Antagonistic effects of resveratrol on reproductive injury in hind-limp unloading male rats[J]. Chinese Journal of Space Science, 2024, 44(1): 133-141 doi: 10.11728/cjss2024.01.2023-0063 [2] ÖZELBAYKAL B, ÖĞRETMENOĞLU G, GEDIK Ş. The effects of space radiation and microgravity on ocular structures[J]. Turkish Journal of Ophthalmology, 2022, 52(1): 57-63 doi: 10.4274/tjo.galenos.2021.29566 [3] GEORGE K, RHONE J, BEITMAN A, et al. Cytogenetic damage in the blood lymphocytes of astronauts: effects of repeat long-duration space missions[J]. Mutation Research/Genetic Toxicology and Environmental Mutagenesis, 2013, 756(1/2): 165-169 [4] MORENO-VILLANUEVA M, WONG M, LU T, et al. Interplay of space radiation and microgravity in DNA damage and DNA damage response[J]. npj Microgravity, 2017, 3(1): 14 doi: 10.1038/s41526-017-0019-7 [5] AFSHINNEKOO E, SCOTT R T, MACKAY M J, et al. Fundamental biological features of spaceflight: advancing the field to enable deep-space exploration[J]. Cell, 2020, 183(5): 1162-1184 doi: 10.1016/j.cell.2020.10.050 [6] DA SILVEIRA W A, FAZELINIA H, ROSENTHAL S B, et al. Comprehensive multi-omics analysis reveals mitochondrial stress as a central biological hub for spaceflight impact[J]. Cell, 2020, 183(5): 1185-1201. e20 [7] LI K, DESAI R, SCOTT R T, et al. Explainable machine learning identifies multi-omics signatures of muscle response to spaceflight in mice[J]. npj Microgravity, 2023, 9(1): 90 doi: 10.1038/s41526-023-00337-5 [8] BEHESHTI A, CHAKRAVARTY K, FOGLE H, et al. Multi-omics analysis of multiple missions to space reveal a theme of lipid dysregulation in mouse liver[J]. Scientific Reports, 2019, 9(1): 19195 doi: 10.1038/s41598-019-55869-2 [9] HOOD L, FLORES M. A personal view on systems medicine and the emergence of proactive P4 medicine: predictive, preventive, personalized and participatory[J]. New Biotechnology, 2012, 29(6): 613-624 doi: 10.1016/j.nbt.2012.03.004 [10] BARABÁSI A L, GULBAHCE N, LOSCALZO J. Network medicine: a network-based approach to human disease[J]. Nature Reviews Genetics, 2011, 12(1): 56-68 doi: 10.1038/nrg2918 [11] HUANG Y H, CHANG X, ZHANG Y, et al. Disease characterization using a partial correlation-based sample-specific network[J]. Briefings in Bioinformatics, 2021, 22(3): bbaa062 doi: 10.1093/bib/bbaa062 [12] ZHANG Y, ZHAO L, SUN Y Q. Using single-sample networks to identify the contrasting patterns of gene interactions and reveal the radiation dose-dependent effects in multiple tissues of spaceflight mice[J]. npj Microgravity, 2024, 10(1): 45 doi: 10.1038/s41526-024-00383-7 [13] ZHANG Y, DU X H, ZHAO L, et al. Construction of dose prediction model and identification of sensitive genes for space radiation based on single-sample networks under spaceflight conditions[J]. International Journal of Radiation Biology, 2024, 100(5): 777-790 doi: 10.1080/09553002.2024.2327393 [14] KUIJJER M L, TUNG M G, YUAN G C, et al. Estimating sample-specific regulatory networks[J]. iScience, 2019, 14: 226-240 doi: 10.1016/j.isci.2019.03.021 [15] GUO W F, YU X T, SHI Q Q, et al. Performance assessment of sample-specific network control methods for bulk and single-cell biological data analysis[J]. PLoS Computational Biology, 2021, 17(5): e1008962 doi: 10.1371/journal.pcbi.1008962 [16] LOVE M I, HUBER W, ANDERS S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2[J]. Genome Biology, 2014, 15(12): 550 doi: 10.1186/s13059-014-0550-8 [17] AN L L, LI Y M, FAN Y J, et al. The trends in global gene expression in mouse embryonic stem cells during spaceflight[J]. Frontiers in Genetics, 2019, 10: 768 doi: 10.3389/fgene.2019.00768 [18] KUMAR A, TAHIMIC C G T, ALMEIDA E A C, et al. Spaceflight modulates the expression of key oxidative stress and cell cycle related genes in heart[J]. International Journal of Molecular Sciences, 2021, 22(16): 9088 doi: 10.3390/ijms22169088 [19] GARRETT-BAKELMAN F E, DARSHI M, GREEN S J, et al. The NASA twins study: a multidimensional analysis of a year-long human spaceflight[J]. Science, 2019, 364(6436): eaau8650 doi: 10.1126/science.aau8650 [20] VAQUER S, CUYÀS E, RABADÁN A, et al. Active transmembrane drug transport in microgravity: a validation study using an ABC transporter model[J]. F1000Research, 2014, 3: 201 doi: 10.12688/f1000research.4909.1 [21] PANDI-PERUMAL S R, GONFALONE A A. Sleep in space as a new medical frontier: the challenge of preserving normal sleep in the abnormal environment of space missions[J]. Sleep Science, 2016, 9(1): 1-4 doi: 10.1016/j.slsci.2016.01.003 [22] MALHAN D, YALÇIN M, SCHOENROCK B, et al. Skeletal muscle gene expression dysregulation in long-term spaceflights and aging is clock-dependent[J]. npj Microgravity, 2023, 9(1): 30 doi: 10.1038/s41526-023-00273-4 [23] FLYNN-EVANS E E, BARGER L K, KUBEY A A, et al. Circadian misalignment affects sleep and medication use before and during spaceflight[J]. npj Microgravity, 2016, 2(1): 15019 doi: 10.1038/npjmgrav.2015.19 [24] CHAKRABORTY N, WANING D L, GAUTAM A, et al. Gene-metabolite network linked to inhibited bioenergetics in association with spaceflight-induced loss of male mouse quadriceps muscle[J]. Journal of Bone and Mineral Research, 2020, 35(10): 2049-2057 doi: 10.1002/jbmr.4102 [25] MAO X W, PECAUT M J, STODIECK L S, et al. Biological and metabolic response in STS-135 space-flown mouse skin[J]. Free Radical Research, 2014, 48(8): 890-897 doi: 10.3109/10715762.2014.920086 [26] CROUCH J D, BROSH JR R M. Mechanistic and biological considerations of oxidatively damaged DNA for helicase-dependent pathways of nucleic acid metabolism[J]. Free Radical Biology and Medicine, 2017, 107: 245-257 doi: 10.1016/j.freeradbiomed.2016.11.022 [27] ROONEY B V, CRUCIAN B E, PIERSON D L, et al. Herpes virus reactivation in astronauts during spaceflight and its application on earth[J]. Frontiers in Microbiology, 2019, 10: 16 doi: 10.3389/fmicb.2019.00016 [28] BRINLEY A A, THERIOT C A, NELMAN-GONZALEZ M, et al. Characterization of Epstein-Barr virus reactivation in a modeled spaceflight system[J]. Journal of Cellular Biochemistry, 2013, 114(3): 616-624 doi: 10.1002/jcb.24403 [29] AGHA N H, MEHTA S K, ROONEY B V, et al. Exercise as a countermeasure for latent viral reactivation during long duration space flight[J]. The FASEB Journal, 2020, 34(2): 2869-2881 doi: 10.1096/fj.201902327R [30] DRAGO-FERRANTE R, DI FIORE R, KAROUIA F, et al. Extraterrestrial gynecology: could spaceflight increase the risk of developing cancer in female astronauts? An updated review[J]. International Journal of Molecular Sciences, 2022, 23(13): 7465 doi: 10.3390/ijms23137465 [31] MORENO-VILLANUEVA M, WU H L. Radiation and microgravity–associated stress factors and carcinogensis[J]. Reach, 2019, 13: 100027 doi: 10.1016/j.reach.2019.100027 [32] BLAKELY E A, KRONENBERG A. Heavy-ion radiobiology: new approaches to delineate mechanisms underlying enhanced biological effectiveness[J]. Radiation Research, 1998, 150(5): S126-S145 doi: 10.2307/3579815 [33] BARCELLOS-HOFF M H, BLAKELY E A, BURMA S, et al. Concepts and challenges in cancer risk prediction for the space radiation environment[J]. Life Sciences in Space Research, 2015, 6: 92-103 doi: 10.1016/j.lssr.2015.07.006 [34] CUCINOTTA F A, TO K, CACAO E. Predictions of space radiation fatality risk for exploration missions[J]. Life Sciences in Space Research, 2017, 13: 1-11 doi: 10.1016/j.lssr.2017.01.005 [35] NANGLE S N, WOLFSON M Y, HARTSOUGH L, et al. The case for biotech on Mars[J]. Nature Biotechnology, 2020, 38(4): 401-407 doi: 10.1038/s41587-020-0485-4 [36] LI Y Q, LIU X Y, LI Y L, et al. USP19 exerts a tumor-promoting role in diffuse large B cell lymphoma through stabilizing PARK7[J]. The FEBS Journal, 2024, 291(21): 4757-4774 doi: 10.1111/febs.17259 [37] LI X, ARSLAN F, REN Y, et al. Metabolic adaptation to a disruption in oxygen supply during myocardial ischemia and reperfusion is underpinned by temporal and quantitative changes in the cardiac proteome[J]. Journal of Proteome Research, 2012, 11(4): 2331-2346 doi: 10.1021/pr201025m [38] ZHANG X M, LUO Y L, LI Q C. TMED3 promotes proliferation and migration in breast cancer cells by activating Wnt/β-catenin signaling[J]. OncoTargets and Therapy, 2020, 13: 5819-5830 doi: 10.2147/OTT.S250766 [39] LIU F, CAO L, ZHANG T, et al. CRL4BRBBP7 targets HUWE1 for ubiquitination and proteasomal degradation[J]. Biochemical and Biophysical Research Communications, 2018, 501(2): 440-447 doi: 10.1016/j.bbrc.2018.05.008 [40] ARJMAND B, REZAEI TAVIRANI M, RAZZAGHI M, et al. Role of Flt4 in skin protection against UVB radiation: a system biology approach[J]. Journal of Lasers in Medical Sciences, 2020, 11(S1): S30-S36 [41] JUNG J J H. SNF1/AMPK-Related Kinase Hunk is Required for Colorectal Tumorigenesis and Inhibits PML-Mediated Suppression of AKT Following DNA Damage[D]. Philadelphia: University of Pennsylvania, 2013 [42] SHARMA N, SPEED M C, ALLEN C P, et al. Distinct roles of structure-specific endonucleases EEPD1 and Metnase in replication stress responses[J]. NAR Cancer, 2020, 2(2): zcaa008 doi: 10.1093/narcan/zcaa008 [43] JAISWAL A S, KIM H S, SCHÄRER O D, et al. EEPD1 promotes repair of oxidatively-stressed replication forks[J]. NAR Cancer, 2023, 5(1): zcac044 doi: 10.1093/narcan/zcac044 [44] SATO R. Characterizing the Role of Nuclear Body Protein ZC3H8 in Proliferation, Apoptosis, and DNA Repair of Mouse Mammary Tumor Cells[D]. Villanova: Villanova University, 2021 [45] RENAUD E, MICCOLI L, ZACAL N, et al. Differential contribution of XPC, RAD23A, RAD23B and CENTRIN 2 to the UV-response in human cells[J]. DNA Repair, 2011, 10(8): 835-847 doi: 10.1016/j.dnarep.2011.05.003 [46] NARDI I K, STARK J M, LARSEN A, et al. USP22 interacts with PALB2 and promotes chemotherapy resistance via homologous recombination of DNA double-strand breaks[J]. Molecular Cancer Research, 2020, 18(3): 424-435 doi: 10.1158/1541-7786.MCR-19-0053 [47] HA K, LEE G E, PALII S S, et al. Rapid and transient recruitment of DNMT1 to DNA double-strand breaks is mediated by its interaction with multiple components of the DNA damage response machinery[J]. Human Molecular Genetics, 2011, 20(1): 126-140 doi: 10.1093/hmg/ddq451 [48] SUN J Y, SU Y J, XU Y N, et al. CD36 deficiency inhibits proliferation by cell cycle control in skeletal muscle cells[J]. Frontiers in Physiology, 2022, 13: 947325 doi: 10.3389/fphys.2022.947325 [49] LIU Y F, QU G Q, LU Y M, et al. Silencing of MAP4K4 by short hairpin RNA suppresses proliferation, induces G1 cell cycle arrest and induces apoptosis in gastric cancer cells[J]. Molecular Medicine Reports, 2016, 13(1): 41-48 doi: 10.3892/mmr.2015.4510 [50] WANG G F, NIU X Y, LIU H N, et al. c-Abl kinase regulates cell proliferation and ionizing radiation-induced G2/M arrest via phosphorylation of FHL2[J]. FEBS Open Bio, 2021, 11(6): 1731-1738 doi: 10.1002/2211-5463.13177 [51] KWON Y, KIM M, JUNG H S, et al. Targeting autophagy for overcoming resistance to anti-EGFR treatments[J]. Cancers, 2019, 11(9): 1374 doi: 10.3390/cancers11091374 [52] HRUSTANOVIC G, LEE B J, BIVONA T G. Mechanisms of resistance to EGFR targeted therapies[J]. Cancer Biology :Times New Roman;">& Therapy, 2013, 14(4): 304-314 [53] LIU S H, JIANG H W, WEN H, et al. Knockdown of tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein zeta (YWHAZ) enhances tumorigenesis both in vivo and in vitro in bladder cancer[J]. Oncology Reports, 2018, 39(5): 2127-2135 [54] LIU Y, LI W, ZHOU S Y, et al. Pan-cancer analysis of the prognostic and immunological role of RPL4[J]. Heliyon, 2024, 10(14): e34461 doi: 10.1016/j.heliyon.2024.e34461 [55] GOEL R K, PACZKOWSKA M, REIMAND J, et al. Phosphoproteomics analysis identifies novel candidate substrates of the nonreceptor tyrosine kinase, Src-related kinase lacking C-terminal regulatory tyrosine and N-terminal myristoylation sites (SRMS)[J]. Molecular :Times New Roman;">& Cellular Proteomics, 2018, 17(5): 925-947 [56] HAESELEER F, SOKAL I, GREGORY F D, et al. Protein phosphatase 2A dephosphorylates CaBP4 and reǵulates CaBP4 function[J]. Investigative Ophthalmology :Times New Roman;">& Visual Science, 2013, 54(2): 1214-1226 -
-





 张彦 男, 1995年7月出生于山东省济宁市, 现为大连海事大学环境科学与工程学院博士研究生, 主要研究方向为空间辐射生物学、计算生物学、人工智能. E-mail:
张彦 男, 1995年7月出生于山东省济宁市, 现为大连海事大学环境科学与工程学院博士研究生, 主要研究方向为空间辐射生物学、计算生物学、人工智能. E-mail: 

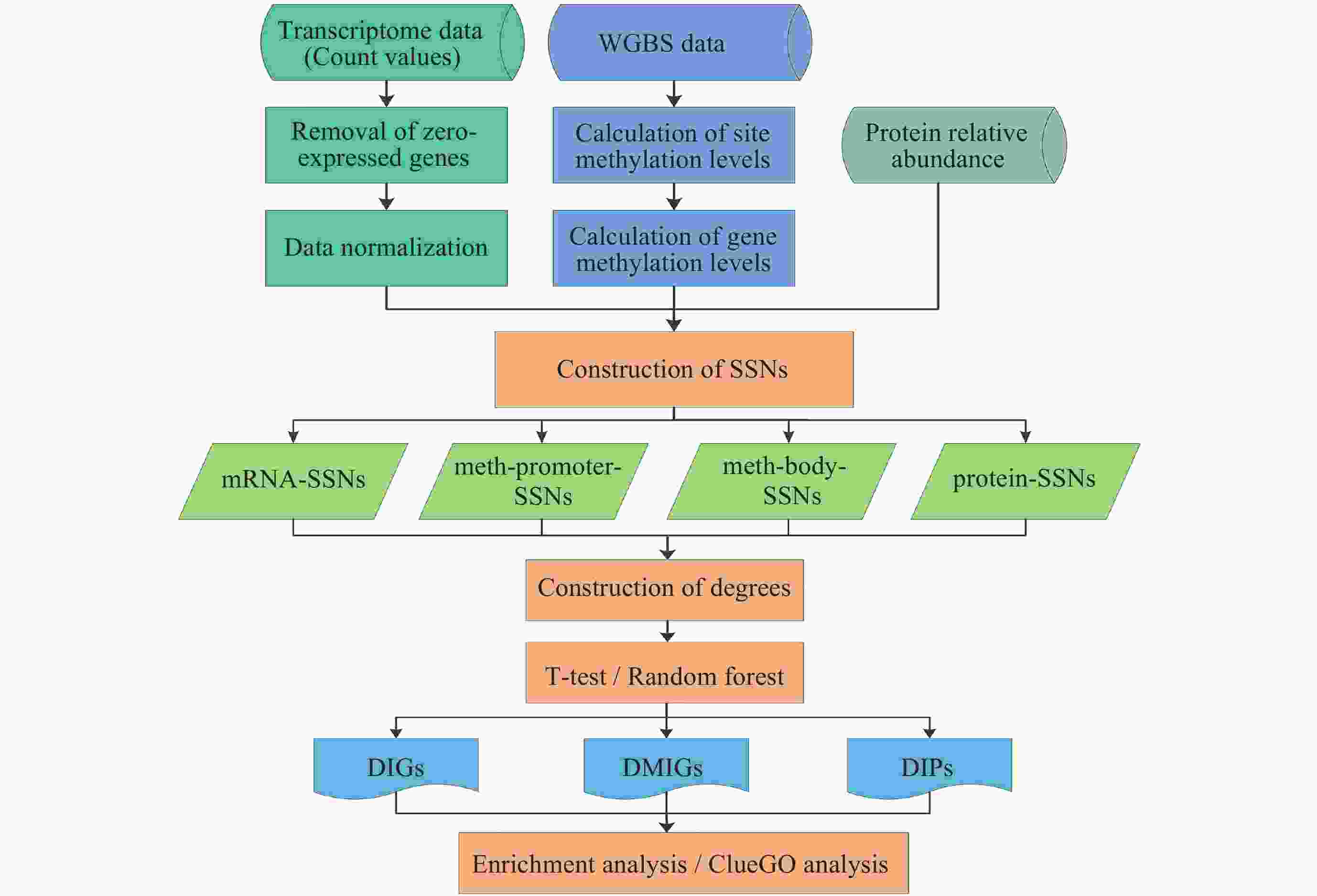
 下载:
下载: